Coronavirus
Baltimore Classification
Higher order taxa
Virus; ssRNA positive-strand viruses, no DNA stage; Nidovirales; Coronaviridae; Coronavirus
Species
Human coronavirus, SARS coronavirus, Bovine coronavirus, Canine coronavirus (examples)
Description and Significance
The coronavirus was first isolated from chickens in 1937. The virus infects not only man but cattle, pigs, rodents, cats, dogs and birds, causing respiratory and enteric diseases. Coronaviruses are the largest positive strand RNA viruses. Coronaviruses cause about one third of common colds and the newly recognized severe acute respiratory syndrome (SARS), making them extremely important.
Genome Structure
The genome of the coronavirus is not segmented and contains a single molecule of linear positive-sense, single-stranded RNA. The complete genome is 27600-31300 nucleotides long. The 5'-end of the genome has a methylated nucleotide cap while the 3'-terminus has a poly (A) tract. (source: ICTV dB Descriptions)
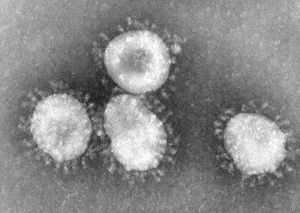
The virions of a coronavirus consist of an envelope and a nucleocapsid that is enveloped. Virions are spherical to pleomorphic and the surface projections are evenly dispersed. They are distinctively clun-shaped peplomers that are spaced widely apart, covering evenly the surface, forming a prominent fringe comprised of SA proteins responsible for attachment to cells, hemagglutination and membrane fusion and hemagglutinin esterase proteins (HE) which form short projections in some species. Different types of proteins make up the 12-24 nm long surface projections. The capsid is elongated with helical symmetry and is 9 nm, or 11-13 nm wide. (source: ICTV dB Descriptions)
The genome replication of coronavirus is mediated through a full-length minus-strand copy of the genome that serves as a template for the production of progeny virus genomes. The transcription of coronavirus is a complex process involving the discontinuous synthesis of up to eight minus-strand RNAs of subgenomic size which contain sequences corresponding to the 5' and 3' ends of the genome and serve as templates for the synthesis of a 5'- and 3'- coterminal set of subgenomic mRNAs. According to the current model of coronavirus discontinuous extension, the synthesis of nascent minus-strand RNA initiates at the 3- end of the plus-strand genomic template and proceeds to TRS elements that are located at various sites within the virus genome. A proportion of the nascent minus-strand RNA is translocated to the 5' leader sequence of the genome, and subsequently a minus-strand copy of the leader is added at these sites. The TRS elements thus determine the fusion sites of leader and "body"-derived sequences of the subgenomic RNAs, and the number of TRS elements correlates with the number of subgenomic mRNAs produced by a particular coronavirus.
The proteins encoded by the replicase gene are critically involved in both replication and transcription. The coronavirus replicase proteins are synthesized as large polyproteins that are extensively processed by virus-encoded proteinases to produce a functional replicase or transcriptase complex. Suggestions have been made that this complex comprises so-called scaffolding proteins, together with proteins that have enzymatic functions, including RNA-dependent RNA polymerase, helicase, and uridylate-specific endoribonuiclease (NendoU), and putative exonuclease and 2'-O-ribose methyl transferase activities. The active replicase or transcriptase complex may contain other proteins of viral or cellular origin in addition.
Viral Ecology & Pathology
Anti-coronaravirus antibodies are present in most people but reinfection is common. This indicates that there are many circulating serotypes of of the virus in the human population. So far, animal reservoirs have not been found for those viruses that infect humans.
Cornavirus causes cold more often in the winter, as is typical with most respiratory infections, because of closer contact. Major outbreaks occur every few years with a cycle that depends on the type of virus that is involved. The human coronaviruses are responsible for 10-20 % of all common colds and have been implicated in gastroenteritis, high and low respiratory tract infections and rare cases of encephalitis.
The symptoms of coronavirus colds are similar to those of rhinovirus colds which include runny nose, sore throat, cough, headache, fever and chills. The incubation period is about 3 days. The immune response of most patients limits the viral spread but this immunity is short-lived. Although it varies considerably between patients, symptoms may last about a week. The patient might shed infectious virus even when there are no apparent symptoms.
Coronaviruses are rather unstable and transmission of the virus is by transfer of nasal secretions caused by sneezes, such as in aerosols. Viruses that infect epithelial cells of the enteric tract cause diarrhea. This is most common in young animals where the infection can be fatal. Although local, coronary infections can spread as well.
Most infections cause a mild, self-limited disease but there may be rare neurological compliations as well. SARS is a form of viral pneumonia where infection encompasses the lower respiratory tract.
Coronaviruses mainly target epithelial cells but they also infect macrophages and other widely distributed cells. These viruses have relatively restricted host ranges, infecting only natural host and closely related animal species. Coronavirus biological vectors are not known.
(source: University of South Carolina: Department of Pathology, Microbiology and Immunology)
References
Luis Enjuanes, Isabel Sola, Fernando Almazan, Javier Ortego, Ander Izeta, Jose M. Gonzalez, Sara Alonso, Jose M. Sanchez, David Escors, Enrique Calvo, Cristina Riquelme, Carlos Sanchez; "Coronavirus derived expression systems"; Journal of Biotechnology 88 (2001) 183–204
Barbara Schelle, Nadja Karl, Burkhard Ludewig, Stuart G. Siddell, and Volker Thiel; "Selective Replication of Coronavirus Genomes That Express Nucleocapsid Protein"; American Society for Microbiology
CDC
The Big Picture Book of Viruses: Coronaviridae
University of South Carolina: Department of Pathology, Microbiology and Immunology