Marivirga tractuosa
A Microbial Biorealm page on the genus Marivirga tractuosa
Classification
Higher order taxa
Domain: Bacteria Phylum: Bacteroidetes Class: Sphingobacteria Order: Sphingobacteriales Family: Flammeovirgaceae Genus: Marivirga
Species
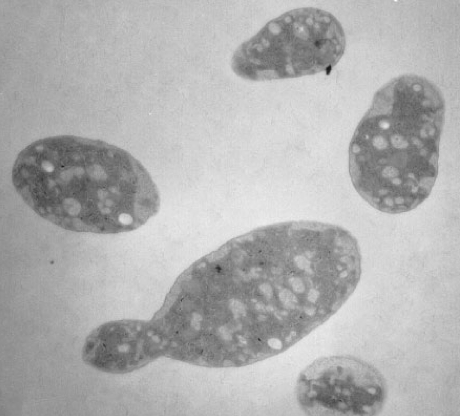
Oceanibulbus indolifex
Description and significance
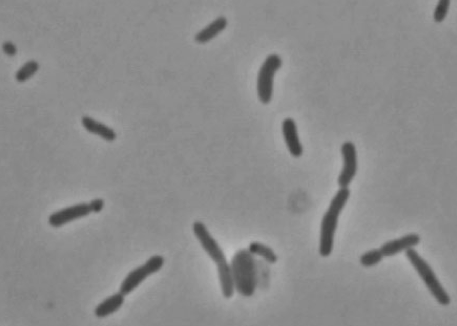
Oceanibulbus indolifex is a rod shaped, gram-negative proteobacteria that forms shiny, white colonies and is not known to be pathogenic to humans. The bacterium was referred to as HEL-45T before it was discovered to be a new species. The bacterium was discovered on September 23, 1998 on the North Sea at 54° 08’ N 7° 52’ E. The sample was taken 2 km from the island of Helgoland at a depth of 10 m. This bacterium is significant because it marks the discovery of a new aerobic species of bacterium that thrives well below the ocean surface.
Genome structure
Oceanibulbus indolifex contains one circular chromosome. The whole genome has not yet been sequence. The complete sequence of the 16S rRNA gene of Oceanibulbus indolifex has a 97.4% identical sequence to Sulfitobacter mediterraneus and a 96.5% identical sequence to Staleya guttiformis. Thus, no other closely related microbes are known, thus supporting the discovery of a new genus and species. Analysis of the genome for Oceanibulbus indolifex indicates 60% G+C content of the DNA.
Cell and colony structure
Oceanibulbus indolifex is a gram-negative bacterium with irregular rod shaped single cells. The rod cells have slightly swollen ends. The cells range in length from 3-5 μm and in width from 1.8-2.5 μm. Oceanibulbus indolifex forms white colored colonies with a characteristic shiny surface. The bacterium consists of white inclusion bodies that are not gas vesicles. These storage granules stain black with Sudan Black stain suggesting they contain poly-β-hydroxybutyrate, a common bacterial storage compound.
Metabolism
Oceanibulbus indolifex is an obligate aerobe, non-fermentative bacterium that requires oxygen to grow. The bacterium is a heterotroph that utilizes D-glucose, pyruvate, DL- lactate, serine, ornithine, alanine, asparagine, L-aspartate, L-glutamate, L-proline, succinate, mannitol, adipate, malate, citrate and glycerol as its carbon source and external electron donor reductant sources. Oceanibulbus indolifex contains Q10 (uibiquinone 10) as its dominant respiratory quinone in the electron transport chain to aid in ATP synthesis. Q10 is specific to members of Alphaproteobacteria class. Oceanibulbus indolifex does not hydrolyze gelatin, starch, urea, or aesculin. The bacterium tested slightly positive for the presence of cytochrome oxidase a transmembrane protein complex in the electron transport chain that transfers electrons to oxygen and translocates four protons per electron to help create a gradient for ATP synthesis. Oceanibulbus indolifex does not reduce nitrate to nitrite. The bacterium produces indole, cyclic dipeptides, and thryptanthrin.
Ecology
Oceanibulbus indolifex has optimum growth from 25-30°C but can grow in a range all the way down to 8°C. The bacterium’s optimum growth occurs at a pH of 7 but it can tolerate a range from 7-9. The optimum salts concentration for growth is 3-5% and the halotolerance range is from 1-10%. Oceanibulbus indolifex does not grow in medium lacking salts or in medium containing only sodium chloride.
Pathology
Oceanibulbus indolifex has no known pathogenic effects on humans. The bacterium does have genes coding for β-lactamases indicating resistance to β-lactam antibiotics like penicillins, cephamycins, and carbapenems. Oceanibulbus indolifex is susceptible to aminoglycosides, antibiotics derived from bacteria of the genus Streptomyces that interfere with bacterial ribosome function.
References
[[3]Wagner-Do ̈bler, I., Rheims, H., Felske, A., El-Ghezal, A., Flade-Schro ̈der, D., Laatsch, H., Lang, S., Pukall, R., Tindall, B., 2004. Oceanibulbus indolifex gen. nov., sp. nov., a North Sea alphaproteobacterium that produces bioactive metabolites. International Journal of Systematic and Evolutionary Microbiology doi: 10.1099/ijs.0.02850-0]
[[4]Piekarski, T., Buchholz, I., Drepper, T., Schobert, M., Wagner-Doebler, I., Tielen, P., Dieter Jahn, D., 2009. Genetic tools for the investigation of Roseobacter clade bacteria. BioMedCentral Microbiology doi:10.1186/1471-2180-9-265]
[[5]Tang, K., Huang, H., Jiao, N., Wu, C., 2010. Phylogenomic Analysis of Marine Roseobacters. PLoS ONE 5(7): e11604. doi:10.1371/journal.pone.0011604]
[[6]Thiel, V., Brinkhoff, T., Dickschat, J., Wickel, S., Grunenberg, J., Simonb, M., Schulz, S., 2009. Identification and biosynthesis of tropone derivatives and sulfur volatiles produced by bacteria of the marine Roseobacter clade. Organic and Biomolecular Chemistry DOI: 10.1039/B909133E]
Edited by Ariel Kaplan of Dr. Lisa R. Moore, University of Southern Maine, Department of Biological Sciences, http://www.usm.maine.edu/bio
