Introduction
Electron micrograph of L. plantarum
By Katie Adlam
At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki. The insertion code consists of:
Double brackets: [[
Filename: PHIL_1181_lores.jpg
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Electron micrograph of the Ebola Zaire virus. This was the first photo ever taken of the virus, on 10/13/1976. By Dr. F.A. Murphy, now at U.C. Davis, then at the CDC.
Closed double brackets: ]]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Lactobacillus plantarum (L. plantarum) is a rod-shaped, gram-positive lactic acid bacterium from the spirochete phylum. It is common found in the human and mammalian gastrointestinal tract and in saliva, as well as in various food products. It can grow at temperatures between 15-45˚C and at pH levels as low as 3.2 (1). L. plantarum is a facultative heterofermentative (2,3), which means that it ferments sugars to produce lactic acid, ethanol or acetic acid, and carbon dioxide under certain conditions and substrates. Depending on the carbon source, these bacteria can switch from using heterofermentative and homofermentative ways of metabolism. L. plantarum is a current interest to researchers and the food industry since it is considered a safe probiotic. This bacterium is acid and bile salt tolerant, which allows it to survive the passage through the gastrointestinal tract of humans.
Genome
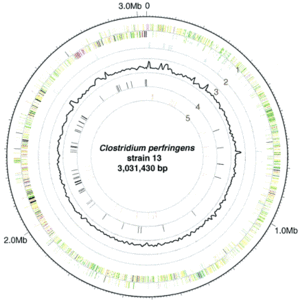
Genome-atlas view of the L. plantarum WCFS1 chromosome, with the predicted origin of replication at the top. The outer to inner circles show (i) positive strand ORFs (red); (ii) negative strand ORFs (blue); (iii) GC-skew (green); (iv) G+C content (black); (v) prophage-related functions (green) and IS-like elements (purple); and (vi) rDNA operons (black) and tRNA encoding genes (red). Kleerebezem 2003.
L. plantarum has a relatively large genome compared to other Lactobacillus spp. Its genome consists of a 3.3 Mb chromosome, which is the largest sequenced genome of any lactic acid bacteria (2). The genome of L. plantarum consists of five rRNA operons, which are evenly distributed around the chromosome (FIGURE, 1). 62 tRNA encoding genes have been found and seem to be in relation to some of the rRNA clusters. In addition, the genome encodes two classes of transposase regions, which are thought to encode mobile genetic elements (1).
The genome consists of 3,052 protein-encoding genes and only 39 of these genes are pseudogenes (1). L. plantarum proteins are very similar to other Gram-positive bacteria since they have low GC content, have a peptidoglycan cell wall, and they are organized collinear. L. plantarum genome contains genes for the Embden-Meyerhoff-Parnas (EMP) pathway and some of the genes correlate to enzymes that break down pentoses and hexoses. Furthermore, its genome shows that it has genes that encode for phosphotransferase, mannose, and fructose systems (1).
Section 2
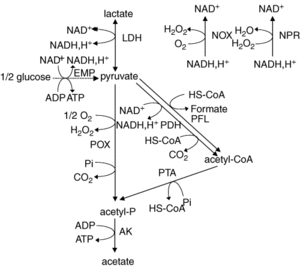
Lactate and acetate production pathways in Lactobacillus plantarum. EMP, Embden–Meyerhof Parnas pathway; LDH, lactate dehydrogenase; POX, pyruvate oxidase; PDH, pyruvate dehydrogenase; PFL, pyruvate formate lyase; PTA, phosphotransacetylase; AK, acetate kinase. Quatravaux 2006.
Include some current research, with at least one figure showing data.
Section 3
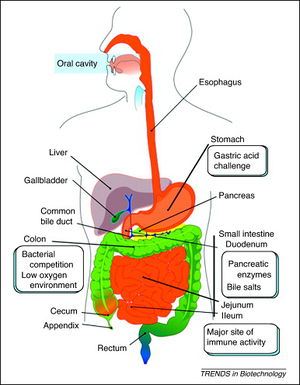
Schematic representation of the human gastrointestinal tract (GI tract) indicating its different regions where recombinant lactic acid bacteria (rLAB) encounter various challenges. Daniel 2011.
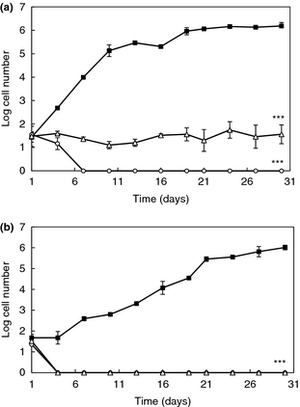
a) Effect of 16 and 62 cCFS on the growth of Rhodotorula mucilaginosa in orange juice. Control juice (), juice containing cCFS from Lactobacillus plantarum 16 (), juice containing cCFS from Lact. plantarum 62 (). P < 0·001 (***). (b) Effect of 16 and 62 cCFS on the growth of R. mucilaginosa in yoghurt. Control yoghurt (), yoghurt containing cCFS from Lact. plantarum 16 (), yoghurt containing cCFS from Lact. plantarum 62 (). P < 0·001 (***). Crowley 2012.
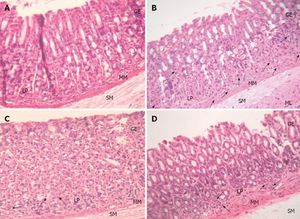
Hematoxylin-eosin stained gastric sections (×20). A: Control group showed normal gastric histopathology; B: Helicobacter pylori infected group showed infiltration of inflammatory cells (arrows); C and D: Lactobacillus plantarum (L. plantarum) B7 106 CFUs/mL treated and L. plantarum B7 1010 CFUs/mL treated groups showed improvements in gastric inflammation. GE: Gastric epithelium; LP: Lamina propria; MM: Muscularis mucosae; SM: Submucosa; ML: Muscularis.Sunanliganon 2012.
Include some current research, with at least one figure showing data.
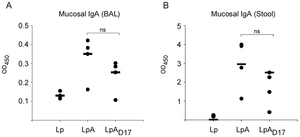
Antibody response to oral administration of recombinant L. plantarum: mucosal IgA. del Rio 2010
Conclusion
Overall text length at least 3,000 words, with at least 3 figures.
References
Kleerebezem, M.; Boekhorst, J.; van Kranenburg, R.; Molenaar, D.; Kuipers, O. P.; Leer, R.; Tarchini, R.; Peters, S.A.; Sandbrink, H.M.; Fiers, M.W.E.J.; Stiekema, W.; Lankhorst, R.M.K.; Bron, P.A.; Hoffer, S.M.; Groot, M.N.N.; Kerkhoven, R.; de Vries, M.; Ursing, B.; de Vos, W.M.; Siezen, R.J. Complete genome sequence of Lactobacillus plantarum WCFS1. PNAS 2003, 100, 1990-1995.
Quatravaux, S.; Remize, F.; Bryckaert, E.; Colavizza, D.; Guzzo, J. Examination of Lactobacillus plantarum lactate metabolism side effects in relation to the modulation of aeration parameters. J Appl. Microbiol. 2006, 101, 903-912.
Daniel, C.; Roussel, Y.; Kleerebezem, M.; Pot, B. Recombinant lactic acid bacteria as mucosal biotherapeutic agents. Trends Biotechnol. 2011, 29, 499- 508.
Crowley, S.; Mahony, J.; van Sinderen, D. Comparative analysis of two antifungal Lactobacillus plantarum isolates and their application as bioprotectants in refrigerated foods. J. Appl. Microbiol. 2012, 113, 1417-1427.
Sunanliganon, C.; Thong-Ngam, D.; Tumwasorn, S.; Klaikeaw, N. Lactobacillus plantarum B7 inhibits Helicobacter pylori growth and attenuates gastric inflammation. World J Gastroenterol. 2012, 18, 2472-2480.
Rio, B.; Seegers, J.F.M.L.; Gomes-Solecki, M. Immune response of Lactobacillus plantarum expressing Borrelia burgdorferi OspA is modulated by the lipid modification of the antigen. PLoS ONE 2010, 5, e11199.
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2011, Kenyon College.







