E. coli and tumor colonization
Introduction
Escherichia coli (Figure 1) is a bacteria commonly found in the intestines of many animals, and it is used as a model organism in many labs.[2] It thrives in a variety of locations due to it being able to respire in both aerobic and anaerobic conditions.[3] E. coli has many diverse strains, with some being pathogenic, harmless, or probiotic.[4] Numerous strains are cultured and grown for research purposes, one such example being E. coli Nissle 1917 (EcN). EcN was isolated in 1917 by Alfred Nissle from the feces of a German soldier. This soldier had been heavily exposed to the pathogenic bacteria Shigella, but did not exhibit any signs of infection from the pathogen. Nissle theorized that the E. coli living in the soldier’s gut had somehow protected the soldier from the Shigella infection. Nissle’s prediction turned out to be correct, as the EcN strain was highly probiotic. EcN was and still is ingested to treat bacterial infections. Genetic analysis of the EcN strain shows that it is entirely non-pathogenic, and that its probiotic effects were due to the production of proteins known as colicins. These colicins inhibit the growth of other bacteria allowing the E. coli to outcompete them. While non-pathogenic, the presence of EcN can still induce a general immune response. This general response can eradicate pathogenic bacteria which increases EcN’s probiotic capabilities.[4]
Cancer and Tumor Colonization
Cancer is a major health concern for the world at large. While many treatment options exist, large solid tumors can prove especially difficult to treat.[5] The central core of the tumor can become hypoxic: deprived of oxygen due to poor vascular organization. The hypoxic region of the tumor grows extremely slowly, limiting the effect of radiation therapies. Combined with a lack of permeability that hinders most drug-therapies, the hypoxic regions of tumors are especially difficult to treat by most methods.[5] A potential solution to this problem was discovered in the late 19th century by William B. Coley.[6] He saw that tumors regressed in patients that had erysipelas infections; this led him to start purposefully infecting patients to treat cancers. This culminated in the creation of “Coley’s Toxins”: mixtures of bacteria designed to treat malignant tumors. While Coley’s initial experiments with live bacteria often led to dramatic tumor regression, they frequently caused dangerous bacterial infections that killed the patient. When he switched to heat-killed bacteria, the danger of deadly infection decreased without hampering the anti-tumor effects.[6]
Coley’s toxins and similar bacterial therapies fell out of use with the advent of other cancer therapies, but with modern techniques failing to treat hypoxic tumors bacterial therapy once again looks promising. Modern research has found that the anaerobic conditions of hypoxic tumors provide an excellent place for anaerobic bacteria to grow.[7] Strintzger et al.[8] found that numerous strains of anaerobic bacteria were able to establish colonies within the hypoxic regions of tumors. EcN was one of the specific strains used, and it successfully colonized the tumor without affecting any other organs.[8] The colonization of tumors by bacteria can alone lead to the regression of the tumor.[9] Similarly to how EcN can induce an immune response against pathogenic bacteria, it is suggested that the colonization of a tumor stimulates an immune response against the tumor. One of the major problems with tumors is that they can evade the immune system by remodeling nearby macrophages to aid the tumor rather than kill it.[10] The accumulation of bacteria within the tumor is able to induce a strong immune response and even “flip” the remodeled macrophages back into tumor suppressors.[11] This immune system stimulation is likely why Coley’s toxins worked even when the bacteria in them were heat killed.[6]
Potentials of Genetic Modification
One of the many reasons for E. coli’s dominance as the model bacteria is its genetic potential.[14] The E. coli species encompasses a wide variety of diverse microbes with a variety of genes. Roughly 80% of all genes sequenced in various E. coli strains are unique to specific strains, having been acquired through horizontal gene transfer. These accessory genes provide advantages for living in a wide range of environments, so the E. coli genome is especially permeable to new genetic information. This makes E. coli a prime candidate for genetic engineering.[14] E. coli can easily be modified to produce various compounds to affect tumors, and this modification has no effect on tumor colonization.[13]
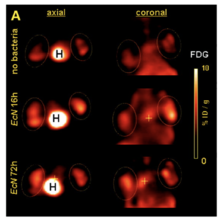
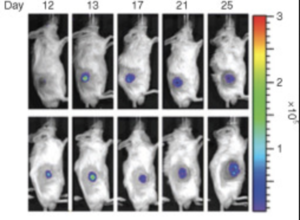
These genetically modified E. coli can be used for tumor imaging. EcN can be genetically modified to produce proteins that fluoresce under UV light. Researchers can then insert these modified bacteria into an organism and measure where they collect or how long they remain in the host.[15] Brader et al., (2008) used genetically modified EcN to colonize and then image tumors. Their EcN produced a fluorescent protein and was grown on radioactive media. After the bacteria were injected into tumor-bearing mice, the size and location of the tumors could be found and displayed. This image could be generated based on the photons emitted by the fluorescent proteins produced by the bacteria (Figure 2). It could also be made from the positron emission from the radioactive sugars ingested by the bacteria (Figure 3).[12]
A promising use of genetically modified bacteria, specifically E. coli Nissle 1917, is the ability to use them as carriers for anti-tumor drugs. It is difficult to deliver drugs using traditional methods to the hypoxic region of large tumors due to a lack of blood vessels.[16] However, anaerobic bacteria such as EcN seek out and colonize these low oxygen hypoxic tumors, which creates a unique transport method right into the tumor. Chiang & Huang[17] made use of engineered EcN as a drug producer and delivery system to combat colorectal cancer. EcN was modified to produce HlyE, a cytotoxic protein capable of lysing mammalian tumor cells. The modified cells were injected into hosts afflicted with cancer. The EcN then colonized the tumors and produced HlyE after the activation of a L-arabinose dependent promoter. The treated tumors decreased in volume by over 75%.[17] In a similar study Zhang et al.[18] used EcN engineered to produce the protein azurine in breast cancer tumors. Once injected into tumor-bearing mice, the EcN quickly accumulated in only the tumor tissue and began producing azurine. The production of azurine in tumor tissue led to the decrease of tumor volume by over 50% compared to the control.[18] Due to findings like these, E. coli Nissle 1917 has strong potential as a treatment for malignant tumors.
References
- ↑ Escherichia coli. Escherichia coli (E. coli). FDA; 2019. Available from: https://www.fda.gov/food/foodborne-pathogens/escherichia-coli-e-coli
- ↑ Blount ZD. The unexhausted potential of E. Coli. eLife. 2015;4.
- ↑ Yasid NA, Rolfe MD, Green J, Williamson MP. Homeostasis of metabolites in Escherichia coli on transition from anaerobic to aerobic conditions and the transient secretion of pyruvate. Royal Society Open Science. 2016Aug1;3(8).
- ↑ 4.0 4.1 Sonnenborn U. Escherichia coli strain Nissle 1917—from bench to bedside and back: History of a special Escherichia coli strain with probiotic properties. FEMS Microbiology Letters. 2016;363(19).
- ↑ 5.0 5.1 Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia. 2015;:83.
- ↑ 6.0 6.1 6.2 Wiemann B, Starnes CO. Coley's toxins, tumor necrosis factor and cancer research: A historical perspective. Pharmacology & Therapeutics. 1994;64(3):529–64.
- ↑ Cao Z, Liu J. Bacteria and bacterial derivatives as drug carriers for cancer therapy. Journal of Controlled Release. 2020Oct10;326:396–407.
- ↑ 8.0 8.1 Stritzker J, Weibel S, Hill PJ, Oelschlaeger TA, Goebel W, Szalay AA. Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. International Journal of Medical Microbiology. 2007Jun11;297(3):151–62.
- ↑ Li R, Helbig L, Fu J, Bian X, Herrmann J, Baumann M, et al. Expressing cytotoxic compounds in Escherichia coli Nissle 1917 for tumor-targeting therapy. Research in Microbiology. 2019Mar;170(2):74–9.
- ↑ Wei B, Pan J, Yuan R, Shao B, Wang Y, Guo X, et al. Polarization of tumor-associated macrophages by nanoparticle-loaded Escherichia coli combined with immunogenic cell death for cancer immunotherapy. Nano Letters. 2021May17;21(10):4231–40.
- ↑ Benoit M, Desnues B, Mege J-L. Macrophage polarization in bacterial infections. The Journal of Immunology. 2008Sep8;181(6):3733–9.
- ↑ 12.0 12.1 Brader P, Stritzker J, Riedl CC, Zanzonico P, Cai S, Burnazi EM, et al. Escherichia coli Nissle 1917 facilitates tumor detection by positron emission tomography and optical imaging. Clinical Cancer Research. 2008Apr15;14(8):2295–302.
- ↑ 13.0 13.1 Jiang S-N, Phan TX, Nam T-K, Nguyen VH, Kim H-S, Bom H-S, et al. Inhibition of tumor growth and metastasis by a combination of escherichia coli–mediated cytolytic therapy and radiotherapy. Molecular Therapy. 2010Mar;18(3):635–42.
- ↑ 14.0 14.1 Blount ZD. The unexhausted potential of E. Coli. eLife. 2015Mar25;4.
- ↑ Schultz M, Watzl S, Oelschlaeger TA, Rath HC, Göttl C, Lehn N, et al. Green fluorescent protein for detection of the probiotic microorganism escherichia coli strain Nissle 1917 (ECN) in vivo. Journal of Microbiological Methods. 2005Jun;61(3):389–98.
- ↑ Taniguchi S, Shimatani Y, Fujimori M. Tumor-targeting therapy using gene-engineered anaerobic-nonpathogenic Bifidobacterium Longum. Methods in Molecular Biology. 2016Feb3;:49–60.
- ↑ 17.0 17.1 Chiang C-J, Huang P-H. Metabolic engineering of probiotic Escherichia coli for cytolytic therapy of tumors. Scientific Reports. 2021Mar12;11(1).
- ↑ 18.0 18.1 Zhang Y, Zhang Y, Xia L, Zhang X, Ding X, Yan F, et al. Escherichia coli Nissle 1917 targets and restrains mouse B16 melanoma and 4T1 breast tumors through expression of Azurin protein. Applied and Environmental Microbiology. 2012Oct5;78(21):7603–10.
Edited by Dominick Frost, student of Joan Slonczewski for BIOL 116 Information in Living Systems, 2022, Kenyon College.
