HIV Drug Resistance
Introduction
By Lillian Spetrino
History of HIV
In 1981, five previously healthy Los Angeles men presented with pneumocystis pneumonia, caused by a fungus that results in the lungs filling with fluid and becoming badly scarred exclusively in people with weakened immune systems. After much dedicated research, it was determined that the cause of these weakened immune systems was a lentivirus termed “HIV” for Human Immunodeficiency Virus. Important research has been conducted to develop drugs that can slow the rise of HIV in infected people, so that onset of AIDs (Acquired Immune Deficiency Syndrome) is delayed (Zimmer).
HIV Structure and Life Cycle
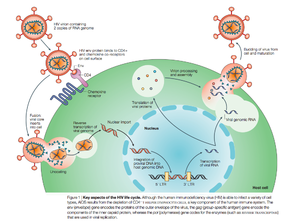
There are two known types of HIV – HIV type 1 and HIV type 2. HIV-2 is less pathogenic than HIV-1 and is thus less widely spread. HIV-1 is a spherical enveloped virus about 120 nm in diameter composed of a protein complex that controls membrane fusion with host CD4+ cells (Chan). The genome of HIV is comprised of two copies of (+) single stranded RNA. HIV infects a variety of immune cells including CD4+ T cells, macrophages, and microglial cells. The transmission of bodily fluids including blood, semen, vaginal fluid, pre-ejaculate, or breast milk transmits infection. Sexual intercourse is the foremost manner of transmission, but is also common through intravenous drug users.
HIV enters the CD4 T cell though tight binding to membrane proteins, causing a conformational change that fuses the membranes and allows entry of the viral capsid in the host cell. Once inside the cell, HIV RNA and reverse transcriptase, integrase, ribonuclease, and protease are used to reverse transcribe the (+) RNA into cDNA, which is transported to the nucleus and into the host cell genome catalyzed by integrase. The virus is then transcribed into mRNA and spliced, exported into the cytoplasm, translated and packaged into virions. These immature virions are transported to the plasma membrane where protease cleaves the gag polyproteins into the matrix, capsid, and nucleocapsid.
HIV Therapy
Prior to 1996, when combination antiretroviral therapy was developed HIV was considered a fatal illness. However, with this development, HIV is now considered a manageable, albeit chronic, disease (Bangsberg). Antiretroviral therapy has decreased HIV-associated morbidity and mortality and changed the disease into a chronic, but controllable illness (Panel on Antiretroviral Guidelines for Adults and Adolescents), as opposed to a death sentence. Antiretroviral therapy is highly effective in transmission prevention between sexual partners (Panel on Antiretroviral Guidelines for Adults and Adolescents). The treatment goals of antiretroviral therapy are to reduce HIV-associated illness and prolong the length and quality of life, repair and preserve immunologic function, suppress plasma HIV viral load, and block HIV transmission (Panel on Antiretroviral Guidelines for Adults and Adolescents). However, an undesired result of antiretroviral therapy can be mutations that lead to decreased drug vulnerability (Novak). Drug-resistance HIV patients have restricted treatment options and have been found to have reduced response to therapy (Novak).
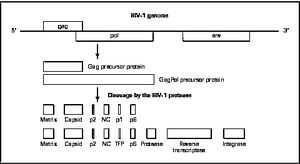
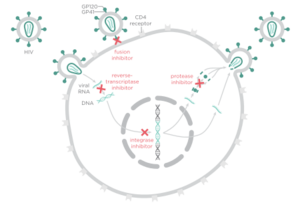
There are four major classes of HIV treatment drugs. One such class is nucleoside and nucleotide analogues. This drug terminates the DNA chain and inhibits reverse transcription of the viral RNA. Another drug class is nonnucleoside reverse-transcriptase inhibitors, which bind and inhibit reverse transcriptase. Additionally, entry inhibitors block the entry of HIV virions into their target cells. Lastly, protease inhibitors target the viral enzyme protease. Protease cleaves the precursor proteins gag and gag-pol, which is important for viral maturation (Clavel). Multiple drug resistance requires ongoing rounds of viral replication in order to create mutations that confer resistance.
Combination therapy is the customary treatment for patients infected with HIV-1. This is the use of three drugs from at least two classes (Panel on Antiretroviral Guidelines for Adults and Adolescents) in order to increase the barrier to resistance. Combination therapy helps to decrease the likelihood of resistance because it reduces HIV replication and thus reduces the likelihood that a resistance mutation will occur. Since resistance requires multiple mutations to develop against all the drugs in the treatment, combination therapy can defend against resistance. However, combination therapy is unable to wholly eliminate resistance to antiretroviral drugs because highly active antiretroviral therapy (HAART) cannot completely overpower viral replication (Smyth).
The goal of HIV treatment is to reduce the amount of virus to indiscernible levels, or less than 50 copies of the virus per mL, ideally 24 weeks post combination therapy. If viral load is found to be higher than 200 copies per mL, further testing must be considered to determine the cause of the virologic failure, which could potentially be resistance. Another way to test how well the treatment is working is to use CD4 cell counts. If the number of CD4 cells is less than 50-100 cells per mL after a year (with 25% fluxuation), it may be indicative of insufficient antiretroviral therapy effectiveness. If there are low viral counts accompanied with low CD4 cell counts, this is termed “immunologic nonresponse” or “immunologic failure” (Panel on Antiretroviral Guidelines for Adults and Adolescents and Gazzola)
Resistance
HIV has a high rate of evolution (Smyth) and therefore abnormally elevated genetic diversity. This genetic variability is a result of the rapid replication cycle. The replication cycle of HIV has a generation time of about 2.5 days, producing between 1010 and 1012 new virions each day. This in combination with the high mutation rate (approximately 0.2 errors by reverse transcriptase per genome during each replication cycle) and the ability to frequently recombine (Rambaut 2004), confers a high mutation rate. High mutation rates can cause drug resistance because modulation of HIV-1 mutation rate by reverse transcriptase can cause polymorphisms that alter replication fidelities resulting in drug resistance (Smyth). Recombination is associated with resistance because drug resistance mutations have been shown to alter the frequency of template switching (Smyth). Template switching is when there is a base-pairing interaction between the 3’ end of a strong-stop DNA and a complementary RNA sequence (Luo) resulting in the production of a recombinant provirus (Bonhoeffer). The genetic diversity in the form of a viral quasispecies is associated with the failure of antiretroviral therapies due to drug resistance (Smyth). Because of the nature of HIV-1 replication and the tendency for rapid selection, high-level drug resistance is a potential hurdle to overcome when treating HIV (Smyth).
Another potential cause of drug resistance in HIV is incomplete adherence. According to the World Health Organization, adherence to long term therapy is “the extent to which a person’s behavior – taking medication, following a diet, and/or executing lifestyle changes, corresponds with agreed recommendations from a health care provider” (World Health Organization). Incomplete adherence therefore is failure to follow such instructions from a health care professional and has often been thought to lead to drug resistance because at low adherence, there is not as much selective pressure to force mutations to arise that may trigger drug resistance. However, recent data has suggested that each regiment class might have a special relationship between adherence and resistance (Bangsberg).
HIV-1 Protease
Structure and Function
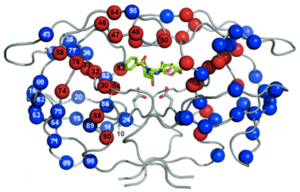
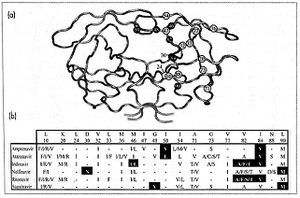
HIV-1 protease is a homodimer composed of two identical subunits of 99 amino acids. Great plasticity and polymorphisms have been observed in 1/3 of the amino acids (Wensing). Also included are two molecular “flaps” that move up to 7 Å when the enzyme is charged with a substrate (Miller). This movement of the flaps is what allows the substrate to enter or depart the cleft (Marseveen). The active site is located between identical subunits and has the classic Asp-Thr-Gly sequence found in many aspartic proteases. More specifically, the catalytic activity of the enzyme is found between the Asp25 residues.
Mature infectious virus particles are generated when HIV protease cleaves viral Gag and GagPol precursor proteins (Wensing). Gag encodes capsid, nucleocapsid, and matrix proteins (Slonczewski), while GagPol encodes RT, integrase, and protease (Slonczewski). Mechanistically, water acts as the nucleophile in combination with aspartic acid to hydrolyze the peptide bond (Jaskolski). This cleavage occurs during or shortly after the assembled viral particles are expelled from host cells and are therefore only essential for viral maturation, but are not required for viral production (Wensing).
Protease Inhibitors
Protease Inhibitor Resistance