Vaginal Microbiota: Canal vs. C-Section
What is the Vaginal Mirobiota?

By Racine Ross
The vaginal microbiota is known to harbor over 250 species of bacteria such as Actinomyces, Bacteroides, Enterobacter, Mollicutes, Proteobacteria, Salmonella, and Streptococcus[1]. In 1892, Albert Döderlein became the first researcher to explain the significance of lactic acid-producing bacteria in the vaginal microbiome [2]. Since then, research has evolved to determine the most prominent species found in the vagina, the most common disturbance of the vaginal microbiome, the varying microbiota types, how the microbiota affects childbirth, and so much more. In normal function, the vaginal microbiota contains a mixture and balance of bacteria such as those previously mentioned. Although, comprehensive surveys of the vaginal microbiome have proved that Lactobacillus species are among the dominant vaginal bacterial species in a large proportion of women [3].
Lactobacillus
Lactobacilli are gram-positive, rod-shaped, non-spore-forming bacteria that grow well in microaerophilic conditions (4). As a gram-positive firmicute, Lactobacilli have a thick cell wall with a cell envelope consisting of glycosyl chains, an s-layer, peptidoglycan with teichoic acids, and a cell membrane containing membrane specific proteins.
Genome of Lactobacillus acidophilus
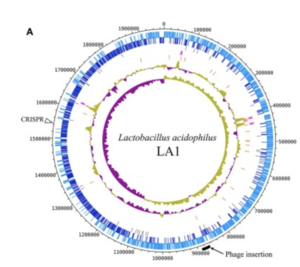
The complete genome is composed of a 1.99-Mbp circular chromosome with 34.7% G+C content, or the percentage of guanine-cytosine in the RNA (Figure1). The genome of Lactobacillus acidophilus was sequenced and a total of 1,953 were found in the genome which included 1,844 protein-coding genes, 76 RNA genes, and 33 pseudogenes. There were 4 sets of ribosomal RNA genes which included 5S, 16S, and 23S genes. Additionally, 61 tRNA genes were found as well as three non-coding RNA.
A unique feature of the Lactobacillus acidophilus is a 26-kbp prophage insertion at the chromosomal position 863,940-890,001. This position and location are similar to other Lactobacillus acidophilus genes, with the only difference being the number of proteins encoded in that specific region (9). LA1, a strain of Lactobacillus acidophilus, contains the smallest number of genes, and the genes required for insertion include attL, phage integrase, and attR (9).
Phylogenetic Comparisons
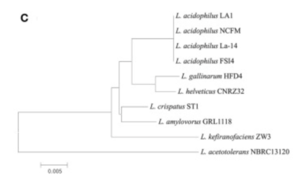
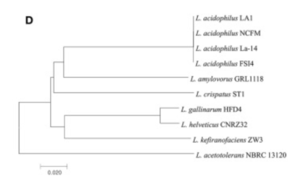
There appeared to be high conservation of the Lactobacillus acidophilus group. The similarity of the genome ranged from 75% to 99.9% for 10 complete genomes in this group. When analyzing similarities based on 16S rRNA, the range was from 94% to 100%. Despite the similarities present, researchers found differing phylogenetic relationships between the 16S rRNA sequence-based and average nucleotide identity value-based phylogenetic trees. Other Lactobacillus species such as Lactobacillus gallinarum and Lactobacillus helveticus were the closest to Lactobacillus acidophilus on the 16S rRNA phylogenetic tree (Figure 2) (9). On the other hand, Lactobacillus amylovorus was the closest to Lactobacillus acidophilus on the average nucleotide identity phylogenetic tree (Figure 3) (9). Studying phylogenetics using average nucleotide identity does a better job at reflecting the functional relationship between strains as opposed to studies that are based on 16S rRNA sequences. Lactobacillus gallinarum and Lactobacillus helveticus showed distinguishable profiles when compared to the Lactobacillus acidophilus genomes (9).
As compared to other species, Lactobacillus acidophilus had a lower G+C content. Logistically speaking, having a lower G+C content would mean that the bonds are less stable as opposed to a species that had a higher G+C content (9). Gene clustering was also completed to understand the difference between 10 genomes that were based on the COG’s database functional annotation. From this database, the genes were categorized into 3,810 functional clusters. A total of 1,717 core gene clusters were found among the total 1,955 gene clusters. Despite the fact that there were high similarities among the Lactobacillus acidophilus genome, four of the gene clusters were found to be absent in the LA1 genome.
Energy Source
Lactobacillus acidophilus was the first probiotic microorganism demonstrated to possess a functional glycogen biosynthetic pathway (5). It is known to be dependent on anaerobic respiration with byproducts including lactic acid from the catabolism of glucose and amino acids. Moreover, Lactobacilli use glucose as a carbon source and can either be homofermentative, meaning that they produce more than 85% of fermentative products as lactic acid, or they can be heterofermentative, meaning that they produce lactic acid, carbon dioxide, ethanol, and acetic acid in equimolar quantities (8).
In its metabolism process, Lactobacillus acidophilus uses the glycogen metabolic pathway. The pathway is encoded by an 11.7-kb chromosomal region that consists of 7 glycogen-specific Lactobacillus acidophilus genes. All seven of these genes are co-transcribed as a polycistronic mRNA and the gene cluster is designated as the glg operon (5). Furthermore, this glg- encoding species primarily originated from mammalian hosts or natural environments.
Researchers have also found that glycogen metabolism is dependent not only on oxygen but also on the carbon source and growth phase. Due to the fact that there is a different expression of the glg operon and glycogen buildup in varying carbohydrate conditions, it shows that glycogen biosynthesis conducted by Lactobacillus acidophilus’’ depends on the type of sugar substrates that are present (5). A microarray study of carbohydrates used by ‘’Lactobacillus acidophilus was conducted which showed that the glg operon was upregulated when raffinose (trisaccharide) or trehalose (a sugar) was given as the sole carbon source as compared to other sugars. Additionally, a PCR experiment, and quantitative glycogen assay showed that raffinose was one of the sugar substrates examined which produces the highest level of the glg operon expression and intracellular glycogen accumulation. On the contrary, the researchers found that glucose appeared to repress the glg expression and biosynthesis. All of this is consistent with the identification of a catabolite response element located upstream of the glg operon. Furthermore, this shows that glycogen metabolism is affected by catabolite regulation (5).
Environmental Interactions
Issues with an imbalance of bacteria
Bacterial Vaginosis
Vulvovaginal Candidiasis
Differences in outcomes between Canal and C-section
Conclusion
References
- ↑ Mendling, W.: Vaginal Microbiota. Advances in experimental medicine and biology, 902, 83–93.
- ↑ Ma, B., Forney, L. J., & Ravel, J. : Vaginal microbiome: rethinking health and disease. Annual review of microbiology, 66, 371–389.
- ↑ Goldstein, E. J., Tyrrell, K. L., & Citron, D. M. (2015). Lactobacillus species: taxonomic complexity and controversial susceptibilities. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America, 60 Suppl 2, S98–S107.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2022, Kenyon College