Naegleria gruberi
Classification
Domain: Eukaryote
Phylum: Protozoa
Class: Lobosa
Order: Amoebida
Family: Vahlkampfidae
Species
Naegleria gruberi
Description and Significance
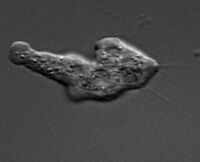
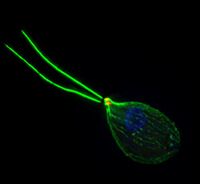
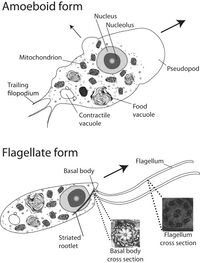
Naegleria gruberi is known as an amoeba-flagellate. This means that it is able to transition from an amoeba to a flagellate and back again. It is approximately 15 microns across and is able to reproduce about every 1.6 hours when feeding on bacteria. N. gruberi can be found in aerobic and microaerobic environments in both fresh water and moist soils around the world [1]. N. gruberi can be seen in its amoeba form in Figure 1 and its flagellate form in Figure 2.
F. Shardinger performed the first recorded transformation of N. gruberi from amoeba to flagellate in 1899. When working in the lab with the amoeba, he found that they kept forming dormant cysts. He reportedly had issues with flagellates forming in the condensation on his agar plates, but chalked it up to contamination. He had attempted to find a developmental stage for the flagellates but they always ended up as cysts. After accidentally innoculating a hanging drop with N. gruberi as amoeba and leaving it for two hours, he came back to discover they had been almost entirely replaced with flagellates [2].
Genome Structure
The various strains of Naegleria gruberi possess varying ploidies. For instance, the strain NEG-M, the only strain which has had its genome fully sequenced is diploid, while the strain NEG is tetraploid. The rest of the information contained in this section pertains to the NEG-M strain. The nuclear Naegleria gruberi genome consists of twelve linear chromosomes. It has a size of 41 Mbp, consisting of at least 15,727 protein-encoding genes. There are two distinct haplotypes present with geometric distributions indicating a history of sexual reproduction. At least 191 of its genes are homologous to genes found in bacteria and archea, with little resemblance to eukaryotic genes.
Outside of the nucleus, each cell possesses 3,000-5,000 copies of a 14 kb circular plasmid which contains all of the rRNA genes for the organism. Furthermore, its mitochondria possess a 50 kb circular genome.
Cell Structure, Metabolism and Life Cycle
In its amoeboid form, N. gruberi has no cytoplasmic microtubule cytoskeleton and the organelles are able to move around within the cell. When transforming into the flagellate form, it creates a cytoplasmic microtubule cytoskeleton from scratch. Two basal bodies and two flagella are also formed. In some cases three or four flagella may be produced, but two is the most common. The basal bodies function to anchor the flagella to the cell and aid in movement. During this phase, the organelles are immobilized. When in the dormant cyst stage, the nucleus size is reduced and the RNA-containing bodies are in the cytoplasms [1]. An image of the amoeboid and flagellate cell structures can be seen in Figure 1.
N. gruberi has three stages of life: amoeba, flagellate, and cyst. During the amoeba phase, its primary stage of life, it feeds on bacteria and reproduces by way of mitosis. In a lab setting, transferring N. gruberi in its amoeba stage from a solid media to a liquid media can cause it to change into a flagellate. Other stressors can cause this change as well, such as temperature, osmotic, or pH changes. After approximately two hours, the flagellate reverts back to an amoeba. When the amoeba reach their stationary phase, they can form resting cysts. This dormant period allows them to survive conditions that would otherwise be fatal to amoeba, such as extreme temperatures, drying, and pH. The cysts are enclosed in a double wall containing several pores. When the time comes to change back, the amoeba crawls out of the cyst through one of the pores [1,3].
Ecology and Pathogenesis
Naegleria gruberi primarily inhabits warm soil and freshwater [4]. Its broad metabolic and physiological capabilities allow it to transition between these differing environments. It preys upon bacteria which it engulfs via phagocytosis [7]. It is unresolved whether Naegleria gruberi undergoes sexual reproduction. It has never been observed under laboratory conditions, despite its genetic geometric distribution indicating that it might. It has been proposed that it only reproduces sexually in the wild, or that it only does so very rarely, and hence nobody has observed it.
Naegleria gruberi is not itself a pathogenic species, however, it is a close relative of the primary amoebic meningoencephalitis causing species Naegleria fowleri. Due to this close relation, it has played a role as a model organism in research related to Naegleria fowleri pathogenesis, both as a safer model organism and as an organism for comparison.
References
[1] Bexkens, M.L., Zimorski,V., Sarink, M.J., Wienk, H., Brouwers, J.F., De Jonckheer, J.F., Martin, W.F., Opperdoes, F.R., van Hellemond, J.J., Tielens, A.G.M. (2018). Lipids are the preferred substrate of the protist maegleriagruberi, relative of a human brain pathogen. Cell Press, 25, 537-543. doi:10.1016
[2] Caprette, D.R. An amoeba-flagellate: naegleria gruberi. Experimental Biosciences. https://www.ruf.rice.edu/~bioslabs/studies/invertebrates/naegleria.html
[3] Fritz-Laylin, L. K., Assaf, Z. J., Chen, S., Cande, W. Z. (2010). Naegleria gruberi de novo basal body assembly occurs via stepwise incorporation of conserved proteins. National Library of Medicine, 9(6), 860-865. Doi: 10.112
[4] Fritz-Laylin, L. K., Prochnik, S. E., Ginger, M. L., Dacks, J. B., Carpenter, M. L., Field, M. C., Kuo, A., Paredez, A., Chapman, J., Pham, J., Shu, S., Neupane, R., Cipriano, M., Mancuso, J., Tu, H., Salamov, A., Lindquist, E., Shapiro, H., & Dawson, S. C. (2010). The genome of Naegleria Gruberi illuminates early eukaryotic versatility. Cell Press, 140:(5), 631-642. doi:10.1016
[5] Fulton, Chandler. (1977). Cell differentiation in naegleria gruberi. Annual Review of Microbiology, 31(1), 597-627. doi:100177.003121
[6] (2003 April 25). Naegleria. Maciver Lab. https://maciverlab.bms.ed.ac.uk/Index.htm
[7] (2010 March). The genome of naegleria gruberi illuminates early eukaryotic versatility. Joint Genome Institute. https://phycocosm.jgi.doe.gov/Naegr1/Naegr1.home.html#:~:text=Naegleria%20gruberi%20
Author
Page authored by Charles Canty and Rachael Bretz, students of Prof. Jay Lennon at IndianaUniversity.