Shewanella frigidimarina
Classification
Bacteria; Pseudomonadota; Gammaproteobacteria; Alteromonadales; Shewanellaceae [Others may be used. Use NCBI link to find]
Species
|
NCBI: [1] |
Shewanella frigidimarina
Description and Significance
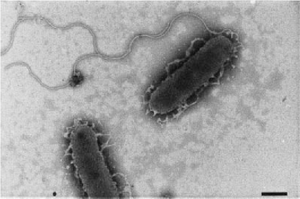
S. frigidimarina is a bacterial species that is motile, gram-negative, and rod-shaped. This psychrophilic species is found in a variety of habitats on the coast of Antarctica (Bozal et al., 2002). It is important to humans because of potential biomedical applications and to the environment because of its use in bioremediation (Sani et al., 2009).
Genome Structure
The genome is made up of about 4.8 million base pairs, with about 41% of that consisting of G & C. This genome exists within a single circular chromosome, although some extrachromosomal plasmids are present. It is very plastic due to the large abundance of mobile genetic elements (MGE) used to evolve and adapt (Di Noto et al., 2016).
Cell Structure, Metabolism and Life Cycle
S. frigidimarina is gram-negative, rod-shaped, and contains one unsheathed polar flagellum for motility. Young colonies are circular and mucoid, while older colonies can sometimes become filamentous. This species is a chemo-organotrophic facultative aerobe, exhibiting the ability to ferment carbohydrayes and reduce iron. When ferric compounds are present, this species produces c-cytochrome to reduce them (Fe(III) to Fe(II)) with DL-lactate as the electron donor (Reyes-Ramirez et al., 2003). It produces polyunsaturated fatty acids through fermentation, that have potential applications in health care and bioremediation of environments affected by petroleum hydrocarbons (due to events like fuel leaks) (Sani et al., 2009).
Ecology
This species inhabits the ice, water, and sediment along the coast of Antarctica. It lives freely in water and sediment as many other bacterial species do, but it produces special ice-binding proteins (IBP) in order to inhabit ice. The ice-binding domains of these proteins protrude from the bacteria to fix it to the surface of the ice (Vance et al., 2018).
References
https://www-webofscience-com.liblink.uncw.edu/wos/woscc/full-record/WOS:000285846200009 https://www-microbiologyresearch-org.liblink.uncw.edu/docserver/fulltext/ijsem/52/1/0520195a.pdf?expires=1668470379&id=id&accname=guest&checksum=76A2EF26D3213D3D9A1F02C7A1E9F8DE https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4859172/
Author
Page authored by Jacob Dehn, student of Prof. Bradley Tolar at UNC Wilmington.