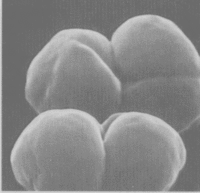
Methanosarcina barkeri
A Microbial Biorealm page on the genus Methanosarcina barkeri
Classification
Higher order taxa
|
NCBI: Archaea : Euryarchaeota : Methanomicrobia : Methanosarcinales : Methanosarcinaceae : Methanosarcina |
Species
|
NCBI: Methanosarcina barkeri |
Strains
Methanosarcina barkeri str. MS
Methanosarcina barkeri str. W
Methanosarcina barkeri str. 227
Description and significance
Methansoarcina barkeri is a large, spherical gram-positive anaerobe that forms packets of varying size. The cells have no flagella and are not mobile.[1] The species have been isolated from lake mud samples, sewage sludge, and is suspected to live in the rumen of the cow. It makes its energy by fermenting small carbon sources, involving the oxidation of H2 and reducing CO2 to methane. The CO2 is produced by pathways starting with methanol, methylamaines, or acetate.
M. barkeri has a thick cell wall (500 nm) and is composed of an acid heteropolysaccharide.
Genome structure
Methanosarcina barkeri fusaro has one circular chromosome and one circular plasmid.
Chromosome
GenBank id: CP000099 Refseq id: NC_007355
Size: 4,837,408 bp (69% coding) 39% GC
3811 genes: 3606 proteins, 73 structural RNAs, 132 pseudogenes
Plasmid
GenBank id: CP000098 Refseq id: NC_007349
Size: 36,358 bp (58% coding) 33% GC
20 genes: 18 proteins, 2 pseudogenes
Cell structure and metabolism
Methanosarcina barkeri is a methanogen and produces methane from a variety of carbon sources, including CO2, acetate, and methanol. It is also an anaerobe and is very oxygen sensitive, so all of its energy production pathways involve some sort of fermentation.
Ecology
Methanosarcina barkeri is an anaerobe has been isolated from mud samples in lakes and bogs. M. barkeri also lives in the rumen of cows where it helps digest organic matter for the cow. A USA Today article has reported that up to 17% of the world's atmospheric methane comes from cows, a large majority of which would come from M. barkeri. Because methane is a green house gas and can interfere with the ozone layer, this small organism may be partially responsible for two of the major environmental crises that we have face: thinning of the ozone layer and global warming.
Pathology
Methanosarcina barkeri is an archaea and therefore causes no known diseases.
Application to Biotechnology
As a methanogen, Methanosarcina barkeri has been looked at as a source of methane (natural gas) for use as an energy source.
Current Research
Longstaff, DG and Blight, SK and Zhang, L and Green-Church, KB and Krzycki, JA. "In vivo contextual requirements for UAG translation as pyrrolysine". Molecular Microbiology. 2007. 63-1, p. 229-241.
Pyrrolysine and selenocysteine are coded by a nonstandard translation of normal stop codons. For selenocysteine, it is known that th insertion of the amino acid instead of message termination is dependent on an insertion sequence that causes the formation of secondary structure in the mRNA that favors insertion of selenocysteine. In this paper, the researchers demonstrate experimentally the requirement of a similar mechanism for pyrrolysine. This sequence, which they call the PYrroLysine Insertion Sequence (PYLIS), is located downstream and is not translated. The experiment consisted of the transfer of a pyrrolysine-containing gene from Methanosarcina barkeri to Methanosarcina acetivorans. This is not a definitive study, as other mechanisms may contribute to the insertion of pyrrolysine or termination.
Sato, T, Atomi, H, and Imanaka, T. "Archaeal Type III RuBisCOs Function in a Pathway for AMP Metabolism". Science. 2007. 315-5817, p. 1003.
Types I and II RuBisCO are found in all phototrophic organisms and are essential enzymes for the carbon fixation of the Calvin cycle. Type IV RuBisCO is found in Bacillus subtilis and is part of the Met salvage pathway. This paper describes the characterization of Type III RuBisCO, which is found only in select archaea, including Methanosarcina barkeri. It was dtermined that Type III RuBisCO catalyzes the reaction of ribulose-1,5-bisphosphate to two molecules 3-phosphoglycerate, which can then enter either glycolysis or gluconeogenesis. This is mediated via two other enzymes (DeoA & E2b2) which break down AMP into the ribulose-1,5-bisphosphate.
Chu, HM and Andrew, HJW. "Enzyme-Substrate Interactions Revealed by the Crystal Structures of the Archaeal Sulfolobus PTP-Fold Phosphatase and its Phosphopeptide Complexes". PROTEINS: Structure, Function, and Bioinformatics. 2007. 66, p. 996-1003.
This paper describes the protein structure of a protein tyrosine phosphatase (PTP) that is important to signal transduction. This is of interest for those interested in Methanosarcina barkeri because protein phosphatases in archaea were first discovered in our favorite bug. This phosphatase was the methyltransferase activation protein, which is important for the activation of the methanol-using pathway of methanogenesis.
Ambrogelly, A, Gundllapalli, S, Herring, S, Polycarpo, C, Frauer, C, and Soll, D. "Pyrrolysine is not hardwired for cotranslational insertion at UAG codons". Proceeds of the National Academy of Sciences. 2007. 104-9, p. 3141-3146.
Pyrrolysine is encoded in Methanosarcinaceae via a naturally encoded tRNA that acts as a termination suppressor. In addition to the tRNA, a pyrrolysine-specific tRNA synthetase must also be present for pyrrolysine incorporation. The researchers in this paper transfered the Pyl tRNA synthetase to Escherichia coli and also transfered different mutants of the tRNA gene. From this, they were able to determine which positions of the tRNA are important for termination suppression based on assays performed. In addition to identifying positions of the tRNA that when mutated reduce suppression, they also found positions that increased suppression.
Feist, AM, Scholten, JCM, Palsson, BO, Brockman, FJ, and Ideker, T. "Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri". Molecular Systems Biology. 2006. vol. 2-1.
The systems biology group at the University of California, San Diego reconstructed the metabolic network of Methanosarcina barkeri. This involved determining reaction kinetics, stoichiometric coefficients, determining metabolic pathways, and determining protein interactions. The reconstruction for Methanosarcina barkeri is especially interesting because it will allow for the simulation of optimized methane-producing mutants as determined by metabolic engineering.
References
Balch, WE, Fox, GE, Magrum, LJ, Woese, CR, and Wolfe, RS. "Methanogens: Reevaluation of a Unique Biological Group". Microbiological Reviews. June 1979. p. 260-296.
Maeder DL, Anderson I, Brettin TS, Bruce DC, Gilna P, Han CS, Lapidus A, Metcalf WW, Saunders E, Tapia R, Sowers KR. "The Methanosarcina barkeri Genome: Comparative Analysis with ...". Journal of Bacteriology. 2006. 188-22, p. 7922-7931.
Stadtman, TC and Barker, HA. "STUDIES ON THE METHANE FERMENTATION IX. The Origin of Methane in the Acetate and Methanol Fermentations by Methanosarcina". Journal of Bacteriology. 1951. 61-1, p. 81-86.
"Cutting cattle's methane emissions". USA Today (Magazine). June 01, 2002.
Atkins, JF and Gesteland, R. "The 22nd Amino Acid". Science. 2002. 296-5572, p. 1409-1410.
US Department of Energy Joint Genome Institute Organism Detail page for Methanosarcina barkeri
TIGR Comprehensive Microbial Resource: Methanosarcina barkeri fusaro Genome Page
Edited by Ian Kerman, a student of Rachel Larsen at UCSD.