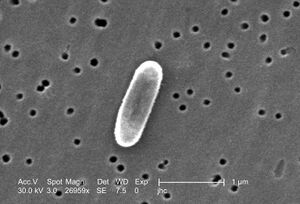
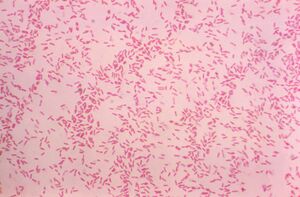
Ralstonia mannitolilytica
Classification
Bacteria; Proteobacteria; Pseudomonadati; Pseudomonadota; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Ralstonia.
Species
|
NCBI: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=105219 |
Ralstonia mannitolilytica
Description and Significance
(Randie) Ralstonia mannitolilytica is a rod-shaped, Gram-negative bacterium found in soil and water. It can survive in many different environments thanks to its ability to use a variety of nutrients and metabolic processes (Ryan et al, 2014) . This makes it useful for environmental cleanup, like breaking down pollutants such as oil spills and other bioremediation. It does so through the lipase it creates, which excels at breaking down the hydrocarbons in oil (Abdulkareem et al, 2024). However, it is also considered an opportunistic pathogen, as it’s known to cause infections in people with weakened immune systems- specifically in hospitals. Its ability to stick to medical devices by forming biofilms and its natural resistance to many antibiotics and other disinfection practices make it difficult to manage in clinical settings. Infections from this are specifically found to be due to long term intravenous medical supplies like catheters (Ryan et al, 2014). Studying this bacterium helps us better understand how to manage its harmful effects while exploring its potential environmental benefits.
Genome Structure
(Elinor) The genome size of reference R. mannitolilytica strain SN82F48 is 5,072,301 bp. It has 2 circular chromosomes and a plasmid. Chromosome 1 is 3,503,899 bp, chromosome 2 is 1,377,860 bp, and plasmid pRMAN01 is 190,542 bp (NCBI, 2015). Carbapenem-resistant strain JARB-RN-0044 has one circular chromosome that is 3.5 Mb and a large replicon plasmid that is 1.5 Mb (Hayashi et al., 2024). Another carbapenem-resistant strain, MRY14-0246, has a total genome size of 4,671,011 bp (Suzuki et al., 2015).
The GC percentage of all three strains are similar: SN82F48 has 65.5% GC, JARB-RN-0044 has 66.1% GC, and MRY14-0246 has 65.8%. Strain SN82F48 has 4472 protein genes and 58 RNA genes, strain JARB-RN-0044 has 3,336 protein-coding genes on the chromosome and 1,390 on the replicon, and strain MRY14-0246 has 4,357 coding sequences (KEGG, 2015; Hayashi et al., 2024; Suzuki et al., 2015). Genome sequencing of JARB-RN-0044 reveals that genes involved in replication, recombination, repair, and metabolism are mostly found on the large chromosome, while genes involved in transcription and cell motility are mostly found on the replicon plasmid (Hayashi et al., 2024).
Novel class D β-lactamase blaOXA-1176 and blaOXA-1177 genes were found in the JARB-RN-0044 strain, while MRY14-0246 was found to have the novel class D β-lactamase genes OXA-443 and OXA-444. These D β-lactamase genes could be a contributing factor to carbapenem class antibiotic resistance in R. mannitolilytica, which makes infections difficult to treat (Hayashi et al., 2024; Suzuki et al., 2015). Furthermore, MRY14-0246 does not have T3SS (Type III Secretion System) gene cluster, but does have the T6SS (Type VI Secretion System). In the plant pathogen Ralstonia solanacearum, T6SS is known to deliver effectors to other organisms that cause cell death, which suggests that T6SS could be a factor in determining the virulence of a Ralstonia species (Suzuki et al., 2015).
Cell Structure, Metabolism and Life Cycle
Ralstonia mannitolilytica is a non-motile, rod-shaped bacilli bacteria with a cell membrane, peptidoglycan within the cell wall, as it is gram-negative, and an outer membrane containing lipopolysaccharides. The bacteria has pili present, which may be a contributing factor to its penchant to form biofilms.
Ralstonia mannitolilytica is a metabolically diverse bacteria. It gains energy through catabolic processes. It oxidizes ammonium to nitrite and can also breakdown carbohydrates like mannitol to gain energy. It is aerobic, but can survive through fermentative processes when oxygen is low or absent.
Because Ralstonia mannitolilytica metabolizes mannitol, it produces polysaccharides Interesting features of cell structure; how it gains energy; what important molecules it produces.
Ecology and Pathogenesis
[This section is in progress]
Habitat; symbiosis; biogeochemical significance; contributions to environment.
If relevant, how does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
References
Abdulkareem, R. S., Al-Ezee, A. M. M., & Musafer, H. K. (2024). New Bioremediation by Lipase Purified from Ralstonia mannitolilytica for Petroleum Hydrocarbons. Samarra Journal of Pure and Applied Science, 6(1), 148–164. https://doi.org/10.54153/sjpas.2024.v6i1.656
Hayashi W, Kaiju H, Kayama S, Yu L, Zuo H, Sugawara Y, Azuma K, Takahashi A, Hata Y, Sugai M. (2024) Complete sequence of carbapenem-resistant Ralstonia mannitolilytica clinical isolate co-producing novel class D β-lactamase OXA-1176 and OXA-1177 in Japan. American Society For Microbiology. 12(4):e03919-23.
Kyoto Encyclopedia of Genes and Genomes. (2015) Ralstonia mannitolilytica. https://www.genome.jp/kegg-bin/show_organism?org=rmn
NCBI. (2015) Ralstonia mannitolilytica genome assembly ASM95413v2. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000954135.1/
Ryan, M. P., & Adley, C. C. (2014). Ralstonia spp.: Emerging global opportunistic pathogens. European Journal of Clinical Microbiology & Infectious Diseases, 33(3), 291-304. SpringerLink.
Suzuki M, Nishio H, Asagoe K, Kida K, Suzuki S, Matsui M, Shibayama K. (2015) Genome sequence of a Carbapenem-resistant strain of Ralstonia mannitolilytica. American Society For Microbiology. 3(3):e00405-15.
Author
Page authored by Clarice Fahrenkrug, Elinor Morkos, Halie Grenier, & Randie Dillon, students of Prof. Bradley Tolar at UNC Wilmington.