Ancient Bacteria and Their Role in Human Evolution and Health
Introduction
The study of ancient bacteria found in archaeological and anthropological contexts has revealed significant insights into human evolution, the microbes that co-evolved with humans, and modern human health. Advances in molecular biology, particularly DNA sequencing and Polymerase Chain Reaction study, have enabled researchers to recover and analyze bacterial DNA from ancient human remains, providing valuable information about past diseases, environmental conditions, and microbial evolution. These studies are revolutionizing our understanding of both human and microbial histories, linking evolutionary events to the microbial world.
Methods of Preservation and Detection
Bacterial Preservation in Ancient Remains
Ancient bacteria have been discovered in many archaeological finds, including among human skeletal remains. These bacteria are often preserved in environments that favor long-term conservation, such as permafrost, dry caves, or sediment layers [1]. The preservation of bacterial DNA in these remains offers an unprecedented opportunity to study the health and microbial life of past populations, as well as the diseases they encountered. This preservation allows scientists to study microbe colonies from over a millennia ago, opening a window into how ancient humans lived and interacted with their environment. Bacteria are preserved in a variety of forms, including living cultures and DNA, allowing for their identification in ancient specimens. [2]. The identification of these bacteria in ancient fossil records illustrates how microbial life can be preserved in archaeological and geological contexts, providing valuable data for studying ancient ecosystems. Researchers use techniques like DNA sequencing and isotopic analysis to uncover traces of ancient microbes in preserved sediments, which is directly relevant to studies of ancient human sites.
Methods of Detection
Advances in molecular biology, especially PCR (Polymerase Chain Reaction) and DNA sequencing, have enabled the identification of ancient bacterial DNA in archaeological and anthropological contexts. These techniques allow researchers to detect and sequence bacterial DNA even when the samples are degraded or fragmented, increasing the ability to study ancient microbiota.[3]. Techniques like these have allowed researchers to identify and sequence bacterial genomes from degraded remains. These methods have opened new doors for the study of ancient microbes, enabling the reconstruction of microbial colonies in the past. The ability to detect bacterial DNA even in extremely degraded samples has transformed how scientists approach ancient health and disease research. [1].
Why Does This Matter?
Human Evolution and Microbial Interactions
Bacteria have been a key factor in shaping human evolution. The discovery of bacterial DNA in ancient human remains helps researchers track the evolution of pathogenic bacteria, immune responses, and the development of human microbiomes. By analyzing the interaction between humans and microorganisms over time, scientists can better understand how ancient microbial communities may have influenced human health and disease resistance. These interactions reveal the dynamic co-evolution of humans and their microbial symbionts, providing insight into how microbial factors may have influenced the course of human history. Some research into ancient microbes suggests that bacterial resistance is not just a product of modern medical practices but a natural evolutionary process that predates human antibiotic use [2].
Health and Disease Evidence
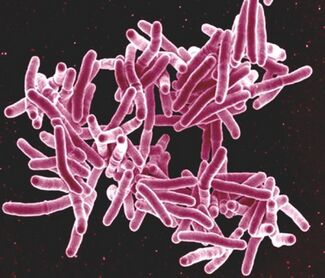
The identification of bacterial DNA in ancient remains has provided crucial evidence for the history of infectious diseases, including tuberculosis (Mycobacterium tuberculosis)[Figure 1.], leprosy, syphilis, and plague (Yersinia pestis)[Figure 2.]. These pathogens are believed to have had significant impacts on ancient populations, influencing human evolution and societal development. For example, the detection of Yersinia pestis in ancient remains has helped scientists understand the spread and impact of the Black Death on medieval populations. This research allows for a deeper understanding of how these diseases shaped the genetics and immunity of modern populations. Understanding how ancient bacteria developed resistance mechanisms can provide valuable context for current issues surrounding antibiotic resistance in modern human populations as well as providing context to human history. [2].
Conclusion
You may have a short concluding section.
Overall, cite at least 5 references under References section.
References
- ↑ 1.0 1.1 “Ancient Bacteria Alive and Well.” Trends in Microbiology, vol. 9, no. 1, Jan. 2001, p. 12, https://doi.org/10.1016/s0966-842x(00)01926-0. Accessed 7 Mar. 2020
- ↑ 2.0 2.1 2.2 “Scientists Find Ancient, Cave-Dwelling Resistant Bacteria | CIDRAP.” Www.cidrap.umn.edu, 12 Dec. 2016, www.cidrap.umn.edu/antimicrobial-stewardship/scientists-find-ancient-cave-dwelling-resistant-bacteria.
- ↑ Johnson SC, Hebsgaard MB, Christensen TR, Mikhail Mastepanov, Nielsen R, Munch K, et al. Ancient bacteria show evidence of DNA repair. 2007 Sep 4;104(36):14401–5.
Edited by [Lily Walker], student of Joan Slonczewski for BIOL 116, 2024, Kenyon College.
