Gloeobacter violaceus
A Microbial Biorealm page on the genus Gloeobacter violaceus
Classification
Higher order taxa
Bacteria; Cyanobacteria; Cyanophyceae; Chroococcales; Gloeobacteraceae
Species
|
NCBI: Taxonomy |
violaceus
Description and significance
It is a rod-shape unicellular cyanobacterium that has been isolated from calcareous rocks in Switzerland [3]. Gloeobacter violaceus is a Gram-negative, photoautotrophic, aquatic and photosynthetic cyanobacteria. Thus Gloeobacter violaceus is commonly labelled as a blue-green algae since it has the ability to live in the water and can manufacture it's own food. Although, it is small and unicellular, Gloeobacter violaceus tend to grow in colonies which are surrounded by a sticky mucous sac that plays a role in adhesion, thus these colonies are large enough to be visible with the human eye [2]. Gloeobacter violaceus is sensitive to strong light and a photoautotroph that contains chlorophyll a, caarotenoids, and phycobiliproteins. Molecular phylogenetic analysis of gloeobacter violaceus has revealed that this linage has diverged from other cyanobacteria and also, that it possesses oxygenic photosynthesis characteristics. Another distinct quality of gloeobacter violaceus is that its' composition of fatty acids is different because it has a high content of polyunsaturated fatty acids (PUFA). A large content of PUFA is unusual for because it was thought that instead there should be a larger composition of (sulfoquinovosyl diacylglycerol SQDG) to ensure photosystem stabilization. These remarkable qualities make it important to sequence the genome of Gloeobacter violaceus because it will reveal the genetic background that is responsible for the origin and evolution of oxygenic photosynthesis. The common method to determine the entire genome of Gloeobacter violaceus is the use of shotgun method in conjunction with the bridging shotgun strategy [3].
Genome structure
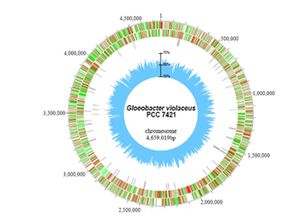
Scientist have revealed that the genome of Gloeobacter violaceus was comprised of a single circular chromosome that contained 4,659,019 base pairs. The average GC content was determined to be 62% and there was also no detection of plasmids during the course of sequencing. The total number of potential protein encoding genes for the entire genome was determined to be 4430 or on average one gene for evey 1052 base pairs. Only 610 genes of gloeobacter violaceus had matches to other cyanobacterial genomes, with about half of these having no known function. Also, about 20 % or 684 genes were found to be unique to this organism [1]. Other interesting features of G. violaceus has been unveiled in comparisons of the assigned gene components with those of other cyanobacteria. It has revealed distinctive features of the G. violaceus genome, such as the genes for PsaI, PsaJ, PsaK, and PsaX for Photosystem I and PsbY, PsbZ and Psb27 for Photosystem II were missing, and those for PsaF, PsbO, PsbU, and PsbV were poorly conserved. CpcG for a rod core linker peptide for phycobilisomes and nblA related to the degradation of phycobilisomes were also missing. These observations can be explained due to the fact that photosynthesis in G. violaceus takes place not in thylakoid membranes but in the cytoplasmic membrane [2].
Cell structure and metabolism
Gloeobacter Violaceus is unicellular organism that exhibits atypical characteristics compared to other cyanobacterium. Its unusual characteristic is the lack of an internal thylakoid membrane system [3]. The rod shaped phycobilisomes of this organism are heavily packed and attach to the plasmic surface of they cytoplasmic membrane. The cell walls of Gloeobacter violaceus are comprised of multiple layers. Starting from the exterior of the cell wall there is a sheath layer, then a double tracked structured outer membrane, intermediate cell wall layer that is electron dense, then lastly, a electron dense peptidoglycan layer. It gets its energy from oxygenic photosynthesis which is unique to this organism because usually the machinery for photosynthesis is found in the thylakoid membrane and not in the cytoplasmic membrane. Important products that G. violaceus produces are petJ and petE which are used for soluble electron transfer catalysts. Also, Gloeobacter violaceus has a lower light requirement for growth than other cyanobacteria, but a higher than normal light saturation level for electron transfer rate [4].
Ecology
The appearance of oxygenic photosynthetic organisms, such as Gloeobacter violaceus, on earth represents the largest discontinuous process in the evolution of photosynthetic organisms. Since the appearance of organisms that are oxygenic photosynthetic, scientist have revealed that they contribute the determining factors that has shifted the direction of global biological evolution through an increase in the oxygen concentration on the Earth [5]. Another important characteristis of the cyanobacterium Gloeobacter violaceus, contains Ths and GroEL and the recombination repair proteins RecA and RadA which enable it to exhibit characterisitics that are similar to Archaea in expressing multiple detoxification PHX bacterioferritin comigratory proteins and several Hsp20s. Also, circadian rhythms have intensively been studied in cyanobacteria, and genes involved in various processes of circadian timing and regulation have been identifiedin many species of cyanobacteria. These genes include kaiABC as the major genetic elements of the circadian clock, sasA, cikA, ldpA, and pex as input modifiers, and rpoD2 and cpmA as output modifiers. Even after scientist completed an intensive search, they could not detect kaiABC in the Gloeobacter genome. It is therefore likely that Gloeobacter does not have a genetic controlling system for circadian rhythms and that cyanobacteria have acquired this system after divergence of the Gloeobacter lineage. Alternatively, Gloeobacter might have lost such genes [2].
Pathology
Gloeobacter violaceus and all species of the genus Gloeobacter are non-pathogenic thus they do not cause any known diseases to humans, animals, or plant hosts.
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used? In cyanobacteria many compounds, including chlorophylls, carotenoids, and hopanoids, are synthesized from the isoprenoid precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate.
Current Research
In a study conducted by Steiger et., al 2005, they revealed that Gloeobacter violaceus is a cyanobacterium isolated from other groups by lack of thylakoids and unique structural features of its photosynthetic protein complexes. They investigated carotenoid biosynthesis with respect to the carotenoids formed and the genes and enzymes involved. Their carotenoid analysis identified ss-carotene as major carotenoid and echinenone as a minor component. This composition is quite unique and the cellular amounts are up to 10-fold lower than in other unicellular cyanobacteria. Also, carotenoid biosynthesis was also found to be up-regulated in a light-dependent manner. This enhanced biosynthesis partially compensates for photooxidation especially of ss-carotene. They also sequenced the genome of G. violaceus and analyzed several gene candidates homologous to carotenogenic genes from other organisms obtained. Functional expression of all candidates and complementation in Escherichia coli led to the identification of all genes involved in the biosynthesis of the G. violaceus carotenoids with the exception of the lycopene cyclase gene. An additional diketolase gene was found that functioned in E. coli but is silent in G. violaceus cells. The biggest difference from all other cyanobacteria is the existence of a single bacterial-type 4-step desaturase instead of the poly cis cyanobacterial desaturation pathway catalyzed by two cyanobacterial-type desaturases and an isomerase. The genes for these three enzymes are absent in G. violaceus.
References
1.Jackisch, Y., Sandmann, G., Steiger, S., "Carotenoid biosynthesis in Gloeobacter violaceus PCC4721 involves a single crtI-type phytoene desaturase instead of typical cyanobacterial enzymes". Archives of Microbiology. 2005. Volume 184
2.Jurgens, U., Schneider, S., "Cell Wall and Sheath Constituents of the cyanobacterium gloeobacter violaceus". Archives of Microbiology. 1991. Volume 155. p. 312-318.
3.Nakamura, Y., Kaneko, T., Sato, S., "Complete Genome Structure of Gloeobacter violaceus PCC 7421, a Cyanobacterium that lacks Thylakoids". DNA Research. 2003. Volume 10. p. 137-145.
4.Rippka, R., "A cyanobacterium which lacks thylakoids." (1974) Arch Microbiol 100, 419-436
5.Mimuro, M., "Photosynthetic properties of a cyanobacterium, Gloeobacter violaceus PCC7421."Department of Physics, Biology and informatics, Faculty of Science, Yamaguchi University. 1-4
6.Steiger, S., "Carotenoid biosynthesis in Gloeobacter violaceus PCC4721 involves a single crtI-type phytoene desaturase instead of typical cyanobacterial enzymes." Arch Microbiology. 2005;184(4):207-14