Halorhodospira halophila
A Microbial Biorealm page on the genus Halorhodospira halophila
Classification
Higher order taxa
Superkingdom: Bacteria;
Phylum: Proteobacteria;
Class: Gammaproteobacteria;
Order: Chromatiales;
Family: Ectothiorhodospiraceae;
Genus: Halorhodospira;
Species: Halorhodospira halophila
Species
Halorhodospira halophila
|
NCBI: Taxonomy |
Strains
Halorhodospira halophila DSM 244/S1
Halorhodospira halophila BN9624
Halorhodospira halophila BN9630
Description and significance
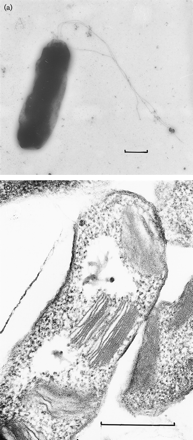
Halorhodospira halophila (formerly Ectothiorhodospira halophila) is an extremely halophilic purple bacterium that was formally a member of the Ectothiorhodospira genus until recently reclassified. Phylogenetically, Halorhodospira halohila is associated within the gamma subdivision of the phylum Proteobacteria and is known to be phototrophic and Gram-negative. It is considered to be “one of the most halophilic eubacteria known.” [2] Its halophilic nature allows for it to be present in conditions that often have been thought to be too harsh for bacteria, and it is found proliferating in saturated salts such as crystallizer ponds (a hypersaline environment where sodium chloride precipitates). [3]
This halophilic organism was first isolated from a salt lake mud [5] has several functions: production of organic solutes glycine, betaine, ectoine, and trehalose to help balance osmotic pressure, as well as the oxidization of sulfide to sulfur (which later would be further oxidized into sulfate). [2]
One important aspect is the production of the bacterial photoreceptor PYP (photoactive yellow protein) within Halorhodospira halophila. PYP is a blue-light sensor found in Halorhodospira halophila that has great significance towards protein research. Photoactive proteins are generally accepted as "model systems for studying protein signal state formation." [4]
Halorhodospira halophila has one major strain (DSM244/SL1) whose genome has been sequenced. But two other strains are recognized: BN9624 and BN9630. [1]
Genome structure
The gene sequence of Halorhodospira halophila S1, the only listed organism of the species has been fully determined. Genome sequencing of Halorhodospira halophila S1 was completed in January 2007 by the Department of Energy Joint Genome Institute. The genome is 2,678,452 nucleotides long (1,339,226 base pairs) and is made up of circular DNA. There are 2493 genes, 2407 which are protein coding, as well as 55 structural RNAs. There is no current information on plasmids related to this species.
Cell structure and metabolism
Halorhodospira halophila is known as a "purple sulfur bacterium", whose structure consists of two membranes as well as the presence of flagella. One important part of Halorhodospira halophila and subject to recent research has been the Photoactive Yellow Protein (PYP), a 14kDa cytoplasmic photoreceptor protein. It generates hydrogen gas during photoautotrophic growth as a byproduct of a nitrogenase reaction.
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
At this time, Halorhodospira halophila causes no known diseases.
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
Enter summaries of the most recent research here--at least three required
Some of the recent research on Halorhodospira halophila includes focusing on it's photobiological ability to produce hydrogen gas and the potential applications of this.
Recent studies of the sequences of the 16sRNA gene have been able to provide details and data into the Ectothiorhodospira genus and the different species within, helping to distinguish Halorhodospira and reassigning it as a separate genus category due to it's extremely halophilic nature.
References
Edited by student Kent Lee of Rachel Larsen and Kit Pogliano
