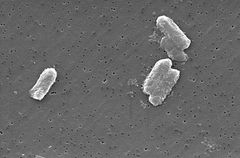
Citrobacter freundii
A Microbial Biorealm page on the genus Citrobacter freundii
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Citrobacter
Species
|
NCBI: Taxonomy |
Citrobacter Freundii
Description and significance
The Citrobacter species, including Citrobacter freundii, are aerobic gram-negative bacilli. Citrobacter freundii is a long rod-shaped bacteria typically 1-5 μm in length. Most have many flagella used to move about, but a few are non-motile. Its habitat includes the enivronment (soil, water, sewage), food, and the intestinal tracts of animals and humans. It belongs to the family of Enterobacteriaceae.
As an opportunistic pathogen, C.freundii is often the cause of significant opportunistic infections. C. Freundii is known to cause a wide variety of nosocomial infections of the respiratory tract, urinary tract, blood and several other normally sterile sites. It represents approximately 29% of all opportunistic infections. Therefore, one of the main reasons the genome of C. Freundii is being sequenced is in order to find antibiotics that can fight these opportunistic infections.
The Citrobacter genus was discovered in 1932 by Werkman and Gillen. Cultures of C. freundii were isolated and identified in the same year from soil extracts.
Genome structure
The complete genome of this microbe has not been sequenced yet since it is so large, although some individual strains and plasmids of the microbe have been sequenced. The most prominent one is the plasmid pCTX-M#3. Its sequence was completed on January 6, 2005. It is a circular DNA plasmid and it is 89,468 nucleotide base pairs long. The length of the plasmid is 0.089468 (Mbp). It is composed of 51.0 % GC content, and encodes 105 proteins.
The C. freundii OS60 AmpC β-lactamase gene has also been sequenced and it is composed of 1197 nucleotides. It encodes a 380 amino acid long precursor with a 19-residue signal peptide. The mature protein encoded by this gene has molecular mass of 39 781 Da. 77% of the amino acid positions hold identical to residues in the E. coli K12 chromosomal AmpC β-lactamases.
Another important feature of the C. Freundii genome is that it is the only microbe in the Enterobacteriaceae family that contains a gene which encodes L_methionine γ_lyase (MGL). The hybrid plasmid pUCmgl obtained from the C. freundii genomic library contains an EcoRI insert which is 3000 base pairs long. The nucleotide sequence of the C. Freundii EcoRI fragment contains two open reading frames. The first frame (the megL gene) encodes a protein of 398 amino acid residues that has sequence homology with MGLs from different sources. The second frame encodes a protein with sequence homology with proteins belonging to the family of permeases.
Another important strain in the genome of C, Freunii GN346, which is a clinical isolate recovered in 1965, and is a typical high producer of a chromosomal claa C beta lactamase. (Haruta) C. Freundii strains also carry a plasmid-encoded AmpC cephalosporinase in its genome that can hydrolyzes and inactivate the anibitoics cephalosporins and cephamycins.
Cell structure and metabolism
The cell structure of C. Freundi is long and rod-shaped usually 1-5 μm in length. The outside of the cell usually contains many flagella used for motality. Since C. freundii is gram-negative bacteria, it contains two membranes (inner and outer). The outer membrane lacks an energy source, but compensates by having porins fused into the membrane. The organism does not have a thick cell wall composed of peptidoglycan like gram-positive bacteria. In between the two membranes lies the periplasmic space. Lipopolysaccharides are anchored to the membrane.
Citrobacter freundii are able to grow on glycerol as the sole carbon and energy source. Glycerol is fermented by a dismutation process involving two pathways. In the first pathway, glycerol is dehydrogenated by a NAD1-linked glycerol dehydrogenase to dihydroxyacetone, which is then phosphorylated and funneled to glycolysis by dihydroxyacetone kinase . In the second pathway, glycerol is dehydrated by the coenzyme B12-dependent glycerol dehydratase to form 3-hydroxypropionaldehyde, which is reduced to the major fermentation product 1,3-propanediol by the NADH-linked 1,3-propanediol dehydrogenase, thereby regenerating NAD1. The four key enzymes of this pathway are encoded by the dha regulon, the expression of which is induced when dihydroxyacetone or glycerol is present
Cells of C. freundii are also able to utilize lactose or citrate as a carbon source.
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
Enter summaries of the most recent research here--at least three required
References
Edited by Sumaira Akbarzada, student of Rachel Larsen