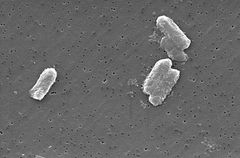
Citrobacter freundii
A Microbial Biorealm page on the genus Citrobacter freundii
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Citrobacter
Species
|
NCBI: Taxonomy |
Citrobacter Freundii
Description and significance
The Citrobacter species, including Citrobacter freundii, are aerobic gram-negative bacilli. Citrobacter freundii are long rod-shaped bacteria typically 1-5 μm in length [1]. Most C. freundii cells are surrounded by many flagella used to move about, but a few are non-motile. Its habitat includes the environment (soil, water, sewage), food, and the intestinal tracts of animals and humans [1]. It belongs to the family of Enterobacteriaceae.
As an opportunistic pathogens, C. freundii is responsible for a number of significant opportunistic infections. It is known to be the cause of a variety of nosocomial infections of the respiratory tract, urinary tract, blood and several other normally sterile sites in patients [2]. C. freundii represents approximately 29% of all opportunistic infections [2]. Therefore, one of the chief reasons many different strains and plasmids of the C. Freundii genome are being sequenced is in order to find antibiotics that can fight these opportunistic infections.
Surprisingly, this infectious microbe in humans plays a positive role in the environment. C. freundii is responsible for reducing nitrate to nitrite in the environment [3]. This crucial conversion is an important stage in the nitrogen cycle. And recycling nitrogen is very essential because the earth's atmosphere is about 85% nitrogen [3]. Therefore, due to its important contribution to the environment is another motivation for sequencing the genome of C. freundii.
The Citrobacter genus was discovered in 1932 by Werkman and Gillen. Cultures of C. freundii were isolated and identified in the same year from soil extracts [1].
Genome structure
The complete genome of this microbe has not been sequenced yet since it is so large, although some individual strains and plasmids of the microbe have been sequenced. The most prominent one is the plasmid pCTX-M#3. Its sequence was completed on January 6, 2005. It is a circular DNA plasmid and it is 89,468 nucleotide base pairs long. The length of the plasmid is 0.089468 (Mbp). It is composed of 51.0 % GC content, and encodes 105 proteins.
The C. freundii OS60 AmpC β-lactamase gene has also been sequenced and it is composed of 1197 nucleotides. It encodes a 380 amino acid long precursor with a 19-residue signal peptide. The mature protein encoded by this gene has molecular mass of 39 781 Da. 77% of the amino acid positions hold identical to residues in the E. coli K12 chromosomal AmpC β-lactamases.
Another important feature of the C. Freundii genome is that it is the only microbe in the Enterobacteriaceae family that contains a gene which encodes L_methionine γ_lyase (MGL). The hybrid plasmid pUCmgl obtained from the C. freundii genomic library contains an EcoRI insert which is 3000 base pairs long. The nucleotide sequence of the C. Freundii EcoRI fragment contains two open reading frames. The first frame (the megL gene) encodes a protein of 398 amino acid residues that has sequence homology with MGLs from different sources. The second frame encodes a protein with sequence homology with proteins belonging to the family of permeases.
Another important strain in the genome of C. freunii is GN346, which is a clinical isolate recovered in 1965, and is a typical high producer of a chromosomal claa C beta lactamase. C. freundii strains also carry a plasmid-encoded AmpC cephalosporinase in its genome that can hydrolyzes and inactivate the anibitoics cephalosporins and cephamycins.
Cell structure and metabolism
The cell structure of C. Freundi is long and rod-shaped usually 1-5 μm in length. The outside of the cell usually contains many flagella used for motality. Since C. freundii is gram-negative bacteria, it contains two membranes (inner and outer). The outer membrane lacks an energy source, but compensates by having porins fused into the membrane. The organism does not have a thick cell wall composed of peptidoglycan like gram-positive bacteria. In between the two membranes lies the periplasmic space. Lipopolysaccharides are anchored to the membrane.
Citrobacter freundii are able to grow on glycerol as the sole carbon and energy source. Glycerol is fermented by a dismutation process involving two pathways. In the first pathway, glycerol is dehydrogenated by a NAD1-linked glycerol dehydrogenase to dihydroxyacetone, which is then phosphorylated and funneled to glycolysis by dihydroxyacetone kinase . In the second pathway, glycerol is dehydrated by the coenzyme B12-dependent glycerol dehydratase to form 3-hydroxypropionaldehyde, which is reduced to the major fermentation product 1,3-propanediol by the NADH-linked 1,3-propanediol dehydrogenase, thereby regenerating NAD1. The four key enzymes of this pathway are encoded by the dha regulon, the expression of which is induced when dihydroxyacetone or glycerol is present
Cells of C. freundii are also able to utilize lactose or citrate as a carbon source.
Ecology
Citrobacter freundii is an enterbacterium commonly found in the environment, mainly in soil, water, and sewages. They are an indicator of potential contamination of water. They are also found on different organs of diseased animals, including mammals, birds, reptiles, and amphibians. They are not known to interact with other organims.
In the environment, C. freundii can convert nitrate or the ammonium ion (which is a nitrogen atom combined with four hydrogen atoms) to nitrite; this reaction occurs in the environment as well as within the digestive tract of humans and other animals. After it converts (reduce) nitrate to nitrite in the environment, the nitrogen cycle is completed when the nitrite is converted to nitrogen. This organism's ecological role not only includes its important role in the nitrogen cycle, it can also accumulate uranium (which is the basic material for nuclear technology) by building phosphate complexes.
Citrobacter freundii has also been investigated for biodegradation of tannic acid used in tannerys.
Pathology
As an opportunistic pathogen, Citrobacter freundii is often the cause of significant opportunistic infections, meaning that it does not generally cause disease in healthy, uncompromised human hosts. Citrobacter species cause a wide variety of nosocomial infections of the respiratory tract, urinary tract, blood and several other normally sterile sites. Hepatic, biliary and pancreatic disease are also common underlying diseases caused by C. Freundii. The biliary tract is the most common site of infection.
One fatal disease that C. freundii has also been associated with is neonatal meningitis and brain abscess. The mortality and morbidity rate of Citrobacter meningitis is unacceptably high. The fatality rate associated with neonatal meningitis is 25 to 50%; moreover, serious neurological sequelae result in 75% of survivors. Although the implication C. freundii in neonatal meningitis and brain abscess is clear, the mechanisms by which these organisms cause disease have been poorly investigated. In this diesease, Citrobacter freundii is able to pentrate blood-brain barrier that consists of the choroid plexus epithelium and the brain capillary endothelium. At this time, it is not known where in the blood-brain barrier C. freundii penetrates. Therefore, there are no cures for this disease.
The diseases observed in rainbow trout and cyprinids were caused by the C. freundii. Histopathological examination confirmed inflammatory changes in the intestine of rainbow trout and inflammatory and necrotic changes in the internal organs of cyprinids. The illness was reproduced by means of artificial infection with a pure culture of C. freundii. This is the first published report confirming C. freundii as a cause of fish disease.
Application to Biotechnology
In the Biotech realm, Citrobacter freundii produces many important enzymes. The first enzymes it produces is phosphatase. It has been reported that strains of Citrobacter freundii and of Staphylococcus aureus accumulated lead as Pb-phosphate when grown on media supplemented with high levels of lead salts. Phosphatase activity, which has been postulated to be involved in lead accumulation, was unrelated to lead resistance, resistant and sensitive cells displaying similar levels and patterns of enzyme activity. Phosphatase activity and lead resistance in Citrobacter freundii
The acid phosphatase of C. freundii has been disocovered to have resistance to some diagnostic reagents. The enzyme of the latter is similar to the PhoN acid phosphatase of Salmonella typhimurium. The ability of a naturally occurring Citrobacter sp. to accumulate cadmium has been attributed to cellular precipitation of CdHPO4, utilizing HPO4(2-) liberated via the activity of an overproduced, Cd- resistant acid-type phosphatase.
C. Freundii strains also carry a plasmid that encodes class 1 AmpC cephalosporinase. These enzymes can hydrolzye inactivate new cephamycins and cephalosporins.
Current Research
A small scale research concerning certain strains of C. freundii was done recently at the University of Tennessee, Knoxville. The importance of certain tetracycline and streptomycin resistance genes and class 1 integrons in C. freundii isolated from dairy farm soil and nondairy soils was evaluated. One strain of C. freundii extracted from dairy farm soils carried class 1 integrons with different inserted gene cassettes. Results of this small study suggest that the presence of multiple resistance genes and class 1 integrons in C. Freundii in dairy farm soil may act as a reservoir of antimicrobial resistance genes and could play a role in the dissemination of these antimicrobial resistance genes to other commensal and indigenous microbial communities in soil. However, additional longer-term studies conducted in more locations are needed to validate this hypothesis.
A second research concering C. freundii was done in order to devise a polymerase chain reaction (PCR) method that simultaneously uses three pairs of specific primers to detect genes of certain microbes (including C. freundii). The method included designing three primer pairs which were: SPVC-1 and SPVC-2, INVA-1 and INVA-2; and VIAB-1 and VIAB-2. PCR was performed using these three primers to identify 14 clinically important bacterial organisms, including Citrobacter freundii, S. enterica serovars Typhi and Paratyphi C, Dublin, and other non-typhoidal Salmonella that harbor a virulence plasmid. The following strains were readily identified using the PCR: (1) C. freundii; (2) S. Typhi; and S. Paratyphi C; (3) S. Dublin (virulence antigen-positive); and (4) Salmonella serovars that harbor an spv-type virulence plasmid. Although this PCR method is new, with the advance of technology in the future this method can allow the identification of C. freundii in mammals immediately so that appropriate antibiotic treatment can be initiated without delay.
A third study concering C.freundii was done in the University of Barcelona, Spain. The mechanisms of resistance to fluoroquinolones in two Citrobacter freundii strains were studied. Both strains were isolated from the same patient and belonging to the same clone by pulsed-field gel electrophoresis. This study allowed partial characterisation of the acrA and acrB genes of this microorganism. The two strains showed the same substitutions in the GyrA and ParC proteins. However, differences were observed in the amount of ciprofloxacin accumulated, with strain 1.38 showing less accumulation. Expression of genes in both strains was analysed using DNA microarrays for Escherichia coli. Nucleotide similarity between the partially sequenced acrA and acrB genes of C. freundii and E. coli was 80.7% and 85%, respectively. The acrA and acrB genes of C. freundii are similar to those in E. coli and their overexpression may play an important role in modulating the final minimum inhibitory concentration of fluoroquinolones in collaboration with mutations in the gyrA and parC genes.
References
1. Wang JT,Chang SC, Chen YC, Luh KT. “Comparison of antimicrobial susceptibility of Citrobacter freundii isolates in two different time periods.” The Journal of Microbiology, Immunology and Infection. 2000 Dec; 33(4): 258-62
2. Whalen JG, Mully TW, Enlgish JC 3rd. “Spontaneous Citrobacter freundii infection in an immunocompetent patient.” Archives of dermatology. 2007 Jan; 143(1): 124-5
3. Puchenkova SG. “Enterobacteria in areas of water along the Crimean Coast.” Mikrobiolohichnyĭ zhurnal. 1996 Mar-Apr; 58(2): 3-7
Edited by Sumaira Akbarzada, student of Rachel Larsen