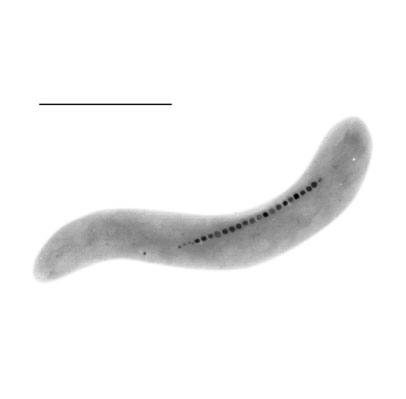
Magnetospirillum magnetotacticum
Classification
Bacteria (Kingdom); Proteobacteria (Phylum); Alphaproteobacteria (Class); Rhodospirillales (Order); Rhodospirillaceae (Class); Magnetospirillum (Family); Magnetospirillum (Genus)
Species
Magnetospirillum magnetotacticum (Synonym: Aquaspirillum magnetotacticum)
Description and Significance
Genome Structure
Magnetospirillum Magnetotacticum has a circular genome, consisting of a total of 9,211,536 base pairs, 66.43% of which are G+C pairs. Protein coding genes comprise 99.08% of the genome, the rest coding for rRNA genes. there are no pseudo genes present in the genome. Though no extrachromosomal structures are found when analyzing undigested DNA, one study found possibile evidence of circular plasmid of 40,000 base pairs that contains 16s rRNA, bra and por genes, made linear and thus detectable by enzyme digestion. An interesting feature of the chromosome is the clustering of genes that contribute this microbes magnetotaxicity. The genes bfr (coding for the iron the iron storage protien bacterioferritin) and magA (coding for the magnetosomal mambrane that surrounds magnetite crystals), are located maximally in the same 17% of the genome, though further studies expect to find that they are indeed much closer together. Also interesting is the overlapping of two bfr genes. Very few magnetotactic bacteria code for two bfr genes, and the few that do, have no overlap. The reason for the overlap is possibly to keep the two genes in proximity, or could play a role in the regulation of the amounts of bacterioferritin protein produced. Magnetospirillum Magnetotacticum was the first magnetotactic bacteria to be phylogenetically analyzed using 16s rRNA genes. This is because it was one of the first magnetotactic bacteria to be isolated in pure culture. Origional analysis placed the bacteria in the aquaspirillum genus, however further studies have placed it in a new genus of magnetospirillum. This study resulted in the folowing evolutionary distance tree.
Cell Structure, Metabolism and Life Cycle
Ecology and Pathogenesis
Magnetospirillum Magnetotacticum was first found by R.P. Blakemore in 1975, isolated from the microarobic zone of a pond. These bacteria, as well as many other magnetotactic bacteria, live in the sediments of fresh water or wet soils. They predominate at the water-sediment interface, an oxygen concentration transition zone, which is consistent with there microaerophillic (sometimes anaerobic) lifestyle. Optimal oxygen conditions for Magnetospirillum magnetotacticum is 3-5% saturation. The environment also has an abundance of dissolved organic acids, utilized by the organism for energy. The main contribution of m. magnetotacticum to the environment is its production of magnetosomes, and the magnetofossiles that are preserved after the death of the organism. These particles of magnetite significantly contribute to the magnetization of soils and sediments. Magnetospirillum magnetotacticum also fixes nitrogen, contributing to the availability of nitrogen in wet soils and sediments. As far as current research goes, Magnetospirillum magnetotacticum has no important symbiotic relationships. This organism is of biogeochemical significance because of its role in iron cycling. It is especially of interest because, understanding the process by which magnetite is formed, along with the overall metabolism can illustrate how the biogeochemical cycles of nitrogen, carbon and iron can be influenced by one organism. Magnetospirillum Magnetotacticum is of geological significance because it leaves a detectible fossil remain, and can be used as a geobiological tracer.
References
1)NCBI: Magnetospirillum magnetotacticum[1]
Author
Page authored by Susan Jarosz and Megan Hull, students of Prof.Jay Lennon at Michigan State University.