Rhodopseudomonas palustris and its role in bioremediation of soil pollutants
Introduction
Rhodopseudomonas palustris belongs to the group of phototrophic non-sulfur purple bacteria. It is a gram negative bacterium commonly found in soil and water. It can survive in both aerobic and anaerobic conditions. It is considered to be a “versatile bacteria” because it is able to fix carbon dioxide to form biomass, convert nitrogen to ammonia and produce hydrogen (hydrogen gas being the byproduct of nitrogen fixation). In the presence of oxygen it catabolizes a range of hydrocarbons from sugars to lignin; however it can switch to photosynthesis in the absence of oxygen. It is increasingly gaining research interest due to its abilities to produce hydrogen and degrade toxic waste. These two have very important applications in bioremediation and alternative energy. [1]
It is in the genes
The
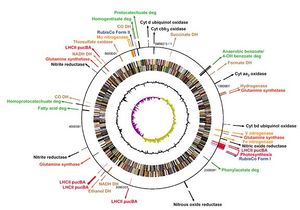
R. palustris genome contains 5,459,213 base-pair circular chromosomes with 4,836 predicted genes and a plasmid of 8,427 bp. 1,514 genes, which is 31% of its genome, code for energy metabolism, biosynthesis, carbon & nitrogen metabolism and cellular processes.
R. palustris possess three nitrogenases, five benzene ring pathways and four light harvesting 2 systems which enable it to carry out its diverse metabolisms. It also has genes that enable it to oxidize hydrogen, thiosulfate and carbon monoxide and use them as energy sources.[2]
Section 2
Include some current research, with at least one figure showing data.
Section 3
Include some current research, with at least one figure showing data.
Conclusion
Overall text length at least 3,000 words, with at least 3 figures.
References
[1]"
Rhodopseudomonas palustris CGA009" Joint Genome Project Institute
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2010, Kenyon College.