Saccharomyces cerevisiae NEU2011
Template:Biorealm Genus Saccharomyces cerevisiae
Classification
Higher order taxa
Domain: Eukarya
Kingdom: Fungi
Subkingdom: Dikarya
Phylum: Ascomycota
Subphylum: Saccharomycotina
Class: Saccharomycetes
Order: Saccharomycetales
Family: Saccharomycetaceae
Genus: Saccharomyces
Species: Cerevisiae
Species
| Taxonomy of Saccharomyces cerevisiae |
Genus species
Description and significance
Saccharomyces cerevisiae is a single-celled Eukaryotic budding yeast belonging to the Ascomycetes, a highly diverse group of fungi. The average cell measures approximately 8 micrometers in diameter, and they typically display a round to ovoid morphology [4]. S. cerevisiae is an important model organism as it was the first Eukaryote to have its genome fully sequenced. (2)
The most important commercial use for S. cerevisiae would be in food production. In the presence of oxygen, yeast cells can reproduce and CO2 by ways of the Citric Acid Cycle. When oxygen is not abundant, S. cerevisiae can also metabolize and reproduce, resulting in CO2 and alcohol by ways of fermentative glycolysis [4]. Used as the primary fermenter in brewing and as a leavening agent in baking, S. cerevisiae ferments sugars into carbon dioxide and alcohol during the brewing and baking processes. Because of these functions, S. cerevisiae is commonly referred to as “Baker’s yeast” or “Brewer’s yeast”. Additionally, the genus name “Saccharomyces” is derived from Latinized Greek and translates to “Sugar Fungus” [2]. The utilization of S. cerevisiae can be dated back to 3150 BC, where strains have been isolated from jars of King Scorpion I 5.
In addition to brewing and breadmaking, Saccharomyces cerevisiae has many pharmaceutical functions. The genome can be engineered to overproduce certain metabolites and commercially important compounds for the use in pharmaceutical agents. Examples of drugs that S. cerevisiae can express compounds for include naringenin, coumarate, artemisinin, taxol, amorphadiene and vitamin C 8.
In nature, S. cerevisiae exists in dense populations where in areas where damaged fruits are located, such as vineyards and fruit farms. S. cerevisiae has additionally been isolated from habitats such as soil, in rivers, and even in the human immune system. It can primarily be found in sugar rich environments, which are ideal for fermentation and reproduction. 3
S. cerevisiae can exist in several different strains. S288c was the first “modern” form of this yeast to be developed in a laboratory setting by Dr. Robert Mortimer the 1950s. As opposed to previously studied strains of yeast, this strain was heterothallic, meaning that the species has different sexes that occur in different individuals, and was therefore adopted as a useful model organism for recombinant research. 3 The S288c strain was later used to isolate other strains of S. cereviae in nature. In order to develop this particular strain, Dr. Mortier performed cross breeding of another heterothallic strain called EM93 that was isolated from rotten figs by Dr. Emil Mrak 3.
Genome structure
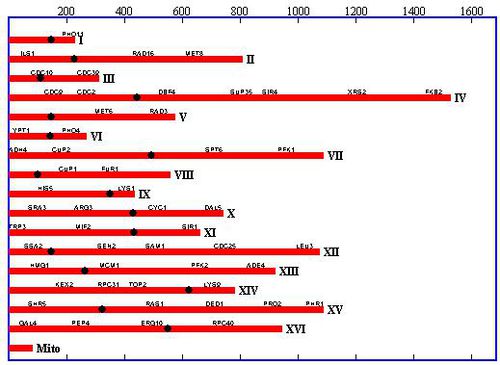
In 1996, Saccromyces cerevisiae was the first Eukaryote to have its genome completely sequenced by a team of over 600 scientists worldwide 5. As of that time, it was the largest genome to be sequenced to date. Because of this, the S. cerevisiae genome has been studied extensively and serves as a model organism for other genomic research. The genome consists of 16 chromosomes (8 pairs) in its haploid vegetative state. In nature, two of these haploid cells will fuse to form a diploid cell with 32 chromosomes (16 pairs). The haploid stage can exist in two forms, α and a, and two cells of the opposite form will fuse to form the diploid cell. S. cerevisiae haploid cells have the ability to switch between the two haploid mating types. The MAT, aka mating type, operon on the S. cerevisiae genome determines which mating type will be expressed. This mating process can produce great genetic variability from generation to generation [4].
When the genome was originally sequenced, it was assumed that all open reading frames longer than 100 codons would code for a protein. In the 15 years to follow, that hypothesis has been continually tested and the genome itself has been updated to contain 522 new ORFs that the originally sequenced genome did not account for 7. On February 2, 2011, an updated release of the S. cerevisiae genome was released 6. This data shows that 70% of the 12,157 kilobase S. cerevisiae genome encodes a protein. This amounts to 4,902 verified open reading frames. In addition to this, 27 genes are known to encode for rRNA, 6 encode for snRNA, and 299 encode for tRNA. This RNA encoding genes are scattered throughout the 16 chromosomes. The updated version of the genome, which can be found at yeastgenome.org, was sequenced using modern Illumina HiSeq, which outdates the Sanger sequencing methods used in 1996. The complete genomes of all major lab strains of S. cerevisiae are expected in the near future using the modern methods.
Cell structure, metabolism & life cycle
Provide a physical and biochemical description of the organism. What kind of organism is it, what does it look like, how is it built, what are its metabolic properties, how can it be identified, what is it's life cycle, &c. In other words, describe the organism from its perspective.
Ecology (including pathogenesis)
Describe its habitat, symbiosis, and contributions to environment. If it is a pathogen, how does this organism cause disease? Human, animal, plant hosts? Describe virulence factors and patient symptoms.
Interesting feature
Describe in detail one particularly interesting aspect of your organism or it's affect on humans or the environment.
References
[2] White, Chris, and Jamil Zainasheff. Yeast: the Practical Guide to Beer Fermentation. Boulder, CO: Brewers Publications, 2010. Print.
[4] Madigan, Michael T. et al, 2009, Brock Biology of Microorganisms, San Francisco, CA, Pearson Education, Inc. Print.
[6] Saccharomyces Genome Database. 18 Feb. 2011. Web. 21 Feb. 2011. <http://www.yeastgenome.org/>.
Edited by the NC State University MB 103 class of 2007.
