Thiobacillus denitrificans
Classification
Bacteria; Protobacteria; Betaprotobacteria; Hydrogenophilales; Hydrogenophilaceae
Species
Thiobacillus denitrificans
Description and Significance
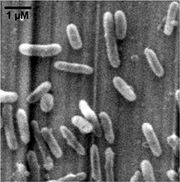
Thiobacillus denitrificans is short rod-shaped (0.5×1.0-3.0 um), Gram-negative, obligately chemolithoautotrophic, and a member of the beta- subclass of the Proteobacteria. Different from many known chemolithotrophic sulfur-oxidizing bacteria (such as Acidithiobacillus ferrooxidans) which are strictly aerobic, T. denitrificans grows as a facultatively anaerobic chemolithotroph, coupling the oxidation of inorganic sulfur compounds to the reduction of oxidized nitrogen compounds (such as nitrate, nitrite) to dinitrogen. The optimum conditions for denitrification are pH 6.85 at 32.8°C, while the optimal growth conditions are pH 6.90 at 29.5°C. This species is widely found in soil, mud, freshwater- and marine sediments, sewage and industrial waste-treatment ponds and digestion tanks which are under anoxic conditions.
T. denitrificans can remediate natural groundwater and engineered water treatment systems by removing excess nitrate. It is the first and only autotrophic bacterium reported to oxidize U(IV) oxide to U(VI) oxide minerals anaerobically, which could partially counteract efforts to remediate uranium contaminated aquifers by in situ reductive immobilization (the radioactive and mobile U(VI) can be dissimilatorily reduced to the immobile form U(IV) by some microorganisms, such as Geobacter sp.).
T. denitrificans is one of the best studied of few obligately chemolithoautotrophic microorganisms which can combine inorganic sulfur-compound oxidation with denitrification.
Genome Structure

The genome of Thiobacillus denitrificans was isolated in 1960s in the soil samples from Texas, USA, and was sequenced by the Department of Energy Joint Genome Institute (DOE JGI).
The strain ATCC 25259 of T. denitrificans has a single, circular chromosome, which is 2,909,809 bp in length, and the DNA base composition consists of an average G+C content of 66.1%. A total of 2886 genes were identified, including 2827 protein-coding genes (97.96%) and 59 RNA genes (2.04%). The region of the 16S rRNA operon (two copies) separates the genome by more than 1,390 kb. It is noticeable that the numbers of its repeated elements, insertion sequence elements (except for a duplicated IS4-like insertion sequence), and transposons are lower than those typically found in bacterial genomes sequenced by far. The biological role categories based on the COG database reveals the largest number of predicted proteins fell into the categories of energy production (6.6%), cell envelope biogenesis (6.0%), and inorganic ion transport (5.7%).
A search using KEGG database of complete genomes revealed to BLAST hits to one of the twelve beta-proteobacteria available in the database. Besides beta-proteobacteria, T. denitrificans also frequently related with Pseudomonas aeruginosa (75 BLAST hits), which is an environmentally versatile gamma-proteobacterium, and interestingly Geobater sulfurreducens (42 hits), which is a delta-proteobacterium best known for its versatility in disimilatory metal reduction (compare to the counteraction mentioned in previous “Description and Significance” section).
Cell Structure, Metabolism and Life Cycle
The cells may be motile through a polar flagellum. Clear or weakly opalescent colonies were observed in the culture under anaerobic condition on thiosulfate/nitrate agar, which might become white with sulfur through time. Nitrogen gas would be produced in anaerobic stab- or roll culture, resulting in agar splitting.
T. denitrificans grows slowly with no apparent stable phase. The maximal denitrification rate reached 2.245 mg/(L-h), which is found in exponential phase. In the process of cultivation, pH of the medium decreases significantly. Some studies reported that relatively high salinity restrained the denitrification activity of T. denitrificans. The acute toxicity test results additionally indicate that T. denitrificans is not toxic.
T. denitrificans grows aerobically on thiosulfate, tetrathionate and thiocyanate; it also grows anaerobically on thiosulfate, tetrathionate, thiocyanate, sulfide or elemental sulfur.
H2S + HS- + NO3- + CO2 + HCO3- + NH4+ --> SO42- + N2 + C5H7O2N (biomass) + H+ + H2O
Previous stoichiometry represents one of the typical reactions describing growth of T. denitrificans on sulfide as an energy source and nitrate as a terminal electron acceptor under the anaerobic condition. Ammonium salts and nitrate are used as nitrogen sources as well. In T. denitrificans, sulfide is oxidized by a series of reactions catalyzed by a sulfide/sulfite oxidoreductase, which is similar to that found in some sulfate-reducing bacteria; and the oxidation of elemental sulfur by T. denitrificans might be preceded by its initial reduction to sulfide, followed by oxidation to sulfite.
In addition, some electron donors that T. denitrificans can utilize are poorly-soluble minerals containing reduced iron and/or sulfur, such as FeS2 and FeS. The mechanism by which this species uses solid-phase electron donors dissimilatorily is of great interest but is yet unknown.
Studies on fixation of carbon dioxide of T. denitrificans found that it is capable of synthesizing hexose phosphates from carbon dioxide by a cyclic mechanism which is similar to that found in green plants. On genomic level, T. denitrificans expresses form I RubisCO under aerobic condition, and form II RubisCO under denitrifying (anaerobic) condition; this discovery not only indicates different mechanisms that influence CO2 fixation efficiencies of T. denitrificans, but also provides some insight into the unusual ability of the species to oxidize sulfur compounds under aerobic and denitrifying conditions.
Batch cultures can be grown in completely filled bottles, resulting in dynamic nitrogen evolution. On the other hand, chemostat culture can be interchanged simply and repeatedly between aerobic and anaerobic growth modes, by repressing/derepressing of nitrate and nitrite reductase synthesis.
Ecology
Thiobacillus denitrificans is a widely distributed bacterium, found in both soil and water habitats. In artificial habitats, Thiobacillus denitrificans is considered easily culturable, and was first cultured by Beijerinck in 1904. The variety of habitats occupied by thiobacillus denitrificans include: pond water, brackish mud, soil, marine sediment, sewage lagoons, digestion tanks, and even abandoned mines. The ideal temperature and pH for growth is 28-32 degrees Celsius and 6.8-7.4, respectively. Although Thiobacillus denitrificans is an anaerobic organism, it can live under aerobic conditions.
There is an interest in Thiobacillus denitrificans for environmental remediation, particularly for nitrate removal. Excess nitrogen can cause problems such as eutrophication and methemoglobinemia (blue baby syndrome), and is caused primarily by discharge of waste waters and overuse of fertilizers. Heterotrophic denitrifiers have traditionally been favored, but Thiobacillus denitrificans may be a good alternative. Thiobacillus denitrificans is an autrophic denitrifier, which has the competitive advantages of not needing an external carbon source to be added to the process, and not producing as much sludge. Sulfur-based systems that T. denitrficans thrive in have been considered as a viable solution to remove nitrate from groundwater and surface water that have been contaminated, as well as remediating waste waters with high nitrate levels such as nitrified leachate. Experimental data supports T. denitrificans as efficient in nitrate removal in the long term, and as a dominant species in nitrate removal in a wide variety of applications. Thiobacillus denitrificans is able to reduce not only nitrate, but nitrite as well.
Thiobacillus denitrificans is also being considered for sulfide removal. Sulfur compounds in the environment are undesirable due to their toxicity, unpleasant smell, corrosive properties, and high oxygen demand. Complete sulfur removal is attained by partial oxidation of sulfide to elemental sulfur followed by phase separation. Aerobic sulfide oxidation is the preferred method for removing sulfide in industrial wastewater, but if both nitrate and sulfide are present, such as is common in the food industry, autotrophic denitrification is favored. The sulfide/nitrate ratio for T. denitrificans may be manipulated control to the fate of sulfide oxidation to either elemental sulfur or sulfate. There is a concern about trace metals and sulfate produced in the sulfide oxidation of Thiobacillus denitrificans, as the oxidation may lead to increased heavy metal motility and toxicity potential, and sulfate may contribute to eutrophication.
In a study of hard coal mining sites in Germany, a high abundance of Thiobacillus denitrificans was found at all sites. In the superficial dump layers of the mining sites, Thiobacillus denitrificans consumed the excess nitrate. Thiobacillus denitrificans was also found in abundance at a constructed wetland for treating mining pollution.
Thiobacillus denitrificans-like (95-96% similarity) organisms have been shown to oxidize U(IV) in the presence of nitrate. These organisms favor growth on the sediment of such contaminated areas, and are associated as playing a large role in the oxidation of reduced minerals. Unfortunately this inhibits remediation of uranium contaminated aquifers by converting the uranium species to a less soluble version.
References
- Kelly, D.P., A.P. Wood, 2000. Confirmation of Thiobacillus denitrificans as a species of the genus Thiobacillus, in the beta-subclass of the Proteobacteria, with strain NCIMB 9548 as the type strain. International Journal of Systematic and Evolutionary Microbiology, 50:547-440
- Shao, M., T. Zhang, H.H. Fang, 2010. Sulfur-driven autotrophic denitrification: diversity, biochemistry, and engineering applications. Applied Microbiology and Biotechnology, 88 (5):1027-1042
- Karyn’s Genomes, European Bioinfomatics Institute.
- Lengeler, J.W., G. Drews, H.G. Schlegel, 1999. Oxidation of Inorganic Compounds by Chemolithotrophs. Biology of the prokaryotes, 10:234-260. [ISBN 3-13-108411-1]
- Genome of Thiobacillus denitrificans ATCC 25259
- Beller, H.R., P.S.G. Chain, T.E. Letain, A. Chakicherla, F.W. Larimer, P.M. Richardson, M. Coleman, A.P. Wood, and D.P. Kelly, 2006. The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans. The Journal of Bacteriology, 188:1473-1488.
- Willscher, S., T. Hertwig, M. Frenzel, M. Felix, and S. Starke, 2010. Results of remediation of hard coal overburden and tailing dumps after a few decades: Insights and conclusions. Hydrometallurgy, 104: 506-517.
- Torrento, C., J. Cama, J. Urmeneta, N. Otero, and A. Soler, 2010. Denitrification of groundwater with pyrite and Thiobacillus denitrificans, 278 (1-2): 80-91.
- N'Guessan, A.L., H.S. Moon, A.D. Peacock, H. Tan, M. Sinha, P.E. Long, and P.R. Jaffe, 2010. Postbiostimulation microbial community structure changes that control the reoxidation of uranium.FEMS Microbiology Ecology, 74 (1): 184-195.
- Moon, H.S., D. Shin, K. Nam, and J.Y. Kim, 2010. Distribution in the Microbial Community Structure in Sulfur-Based Autotrophic Denitrification Columns. Journal of Environmental Engineering, 136 (5): 481-486.
- Thiobacillus denitrificans (strain ATCC 25259), UniProt Consortium
Authors
Page authored by Emily Campbell and Rui Chen, students of Prof. Jay Lennon at Michigan State University.
