Chromosomes in Bacteria: Are they all single and circular?
Introduction
For many years, all bacteria were believed to have a single, circular chromosome in comparison to the eukaryotes, which have linear chromosomes. Perhaps, this was due to the limited sample of bacterial strains studied at the time and the sequencing techniques available to examine the bacterial chromosomes. However, this belief was disproved when bacteria with multiple and/or linear chromosomes were discovered in 1989.
Historical Origin
Single and Circular Chromosome
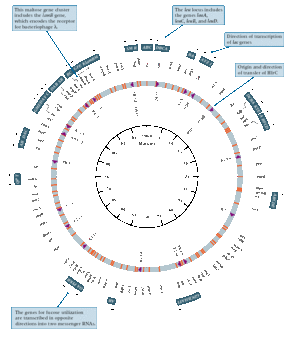
In 1963, studies conducted by several researchers determined that the Escherichia coli genome was organized into a single, circular chromosome [4]. The evidence for the structure of chromosomal DNA was shown by images achieved through autoradiography, electron microscopy and moving pictures of DNA using fluorescence microscopy. Cairns was the first researcher to obtain an image of the entire chromosome for E. coli [4]. The technique used was autoradiography where the chromosome for E. coli was labeled using tritium labeled thymidine, a radioactive isotope of hydrogen [4]. However, the sizes of the chromosomes were variable and there was a low frequency of circular forms detected. Further experimentation based on Hfr conjugation convincingly demonstrating that E. coli has a circular chromosome [3].
The published data of the circular chromosome in E. coli was quickly adopted by many researchers. Therefore, E. coli became widely used as the primary model to investigate chromosomal replication [6].
Multiple Chromosomes
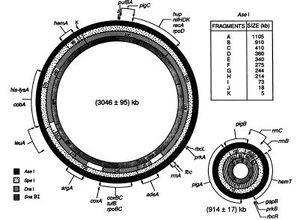
The first evidence of multiple chromosomes in bacteria was found in Rhodobacter sphaeroides, a rod-shaped, Gram-negative bacterium (Figure 2) [12]. The researchers were able to provide a complete physical map of the R. sphaeroides genome by obtaining chromosomal DNA fragments through restriction digest with AseI, SpeI, DraI, and SnaBI from the genomic DNA, which aided in proving the existence of multiple chromosomes [12]. The pulse-field get electrophoresis (PFGE) is used to separate the DNA molecules by applying an electric field. Unlike the standard gel electrophoresis, the voltage is constantly switched in three different directions [3]. The assumption here is that circular DNA is immobile while most of the DNA is mobilized by breakage [3]. The advantage of the PFGE is that it can separate DNAs from a few kilobases (kb) to over 10 megabase (Mb) pairs. The DNA fragments obtained from using endonucleases such as SnaBI produce a distinct pattern of bands useful for physical mapping of the chromosomes. The first evidence leading to the idea of two circular chromosomes was from genomic DNA digestion using SnaBI and AseI, where the fragments were separated using PFGE and hybridization with hemA as a probe [12]. The results showed that the SnaBI fragment of 784 kb was located entirety within the AseI fragments D, E, and H of 914 kb [12]. However, there is a 130 kb of DNA unaccounted, which makes it arithmetically impossible since there must be a total SnaBI fragment length of 1,225 kb [12]. Therefore, the AseI fragments D, E and H cannot be joined with the other contiguous AseI fragments [12]. Through the process of narrowing down how the fragments connect, the chromosomes were physically mapped. In a molecular-basis, this experiment was used to determine that R. sphaeroides have two circular chromosomes, where one is 3.1 Mb and the other is 0.9 Mb [12]. In terms of a genetic-basis, the interrupted mating experiments using Hfr strains strengthened the evidence for the presence of multiple chromosomes in R. sphaeroides [11].
Linear Chromosomes
In 1970, the first evidence of linear chromosomes was discovered, but the limited availability of scientific techniques, and a strong belief of bacterial chromosomal circularity was so convincing that many people did not take in this idea till much later. By 1989, the pulse-field gel electrophoresis technique was developed, and used in conjunction with restriction digests to verify that spirochete Borrelia burgdorferi has a linear chromosome using a similar process of narrowing down by connecting the separated fragments like the R. sphaeroides [2]. B. burgdorferi was the first bacterium to show that linear chromosomes are present in prokaryotes, and the size of the chromosome was found to be around 1.0 Mb [2]. This chromosome organization is comparable to the eukaryotic chromosome organization.
Problems with Linear Chromosomes
Two problems arise with the presence and use of linear chromosomes compared to circular chromosomes in prokaryotes. Firstly, the free double-stranded DNA ends must have some kind of protection so that intracellular nucleases do not degrade them [14]. Secondly, the telomeres, which are the ends of the linear DNA molecules, will require a different process for DNA replication [14]. For protection, there are two types of telomeres that have been observed. The first one is called the palindromic hairpin loops, in which there are no free double-stranded ends available [8], [14]. The other type is invertron telomeres where the chromosomes contain a protein that binds to the 5’-ends [8], [14]. This can be found in Streptomyces linear chromosomes. In terms of DNA replication of the ends of the linear DNA molecules, not much is known about the mechanism.
Other Findings of Linear Chromosomes
Streptomyces griseus
S. griseus is a Gram-positive bacterium known to produce the antibiotic, streptomycin [10]. This bacterium also has a linear chromosome identified through pulse-field gel electrophoresis in conjunction with restriction digests [10]. There are also other Streptomyces species with linear chromosomes identified.
Agrobacterium tumefaciens
Using pulse-field gel electrophoresis, the A. tumefaciens genome was analyzed, where one linear chromosome and one circular chromosome were identified [1]. The size of the linear chromosome is 2100 Kb, which is smaller than the 3000 Kb circular chromosome [1]. The Agrobacterium species have a close 16s rRNA sequence relationship with Brucella species, which have members known to have two circular chromosomes [1].
Other Findings of Multiple Chromosomes
Brucella melitensis
B. melitensis is a Gram-negative coccobacillus bacterium, which has two circular chromosomes [5]. The two chromosome sizes are 2117 Kb and 1178 Kb [5]. Other Brucella species with two circular chromosomes include B. suis biovar 1, B. suis biovar 2, B. suis biovar 4, B. abortus and B. ovis [9].
Paracoccus denitrificans
P. denitrificans is a special bacterium with three circular chromosomes identified by pulse-field gel electrophoresis [15]. The approximate sizes of the chromosomes are 2, 1.1, and 0.64 Mb [15].
Vibrio Species
There are many other bacteria with multiple chromosomes, but Vibrio species has members that have two circular chromosomes [16]. These include V. cholerae, V. parahaemolyticus, V. vulnificus and V. fluvialis [16].
Summary of Currently Known Bacterial Chromosome Organizations
| Bacteria | Chromosome Organization |
|---|---|
| Agrobacterium tumefaciens | One linear and one circular |
| Bacillus subtilis | Single and circular |
| Bacillus subtilis | Single and linear |
| Borrelia burgdorferi | Two circular |
| Brucella abortus | Two circular |
| Brucella melitensis | Two circular |
| Brucella ovis | Two circular |
| Brucella suis biovar 1 | Two circular |
| Brucella suis biovar 2 | Two circular |
| Brucella suis biovar 4 | Two circular |
| Escherichia coli | Single and circular |
| Paracoccus denitrificans | Three circular |
| Pseudomonas aeruginosa | Single and circular |
| Rhodobacter sphaeroides | Two circular |
| Streptomyces griseus | Linear |
| Vibrio cholerae | Two circular |
| Vibrio fluvialis | Two circular |
| Vibrio parahaemolyticus | Two circular |
| Vibrio vulnificus | Two circular |
References
(1) Allardet-Servent, A., Michaux-Charachon, S., Jumas-Bilak, E., Karayan, L. and Ramuz, M. “Presence of One Linear and One Circular Chromosome in the Agrobacterium tumefaciens C58 Genome.” Journal of Bacteriology, 1993, 175(24): 7869-7874
(2) Baril, C., Richaud, C., Baranton, G. and Saint-Girons, I. “Linear Chromosome of Borrelia burgdorferi.” Res. Microbiol., 1989, 140: 507-516
(3) Bendich, A.J. “The Form of Chromosomal DNA Molecules in Bacterial Cells.” Biochimie, 2001, 83: 177-186
(4) Cairns, J. “The Bacterial Chromosome and its Manner of Replication As Seen By Autoradiography.” J. Mol. Biol., 1963, 6: 208-213
(5) Del Vecchio, V.G., Kapatral, V., Elzer, P., Patra, G. and Mujer, C.V. “The Genome of Brucella melitensis.” Veterinary microbiology, 2002, 90: 587-592
(6) Egan, E.S., Fogel, M.A. and Waldor, M.K. “Divided Genomes: Negotiating the Cell Cycle in Prokaryotes with Multiple Chromosomes.” Molecular Microbiology, 2005, 56(5): 1129-1138
(7) Griswold, A. “Genome Packaging in Prokaryotes: the Circular Chromosome of E. coli.” Nature Education, 2008, 1(1)
(8) Hinnebusch, J. and Tilly, K. “Linear Plasmids and Chromosomes in Bacteria.” Molecular Microbiology, 1993, 10(5): 917-922
(9) Jumas-Bilak, E., Michaux-Charachon, S., Bourg, G., O’Callaghan, D. and Ramuz, M. “Differences in Chromosome Number and Genome Rearrangements in the Genus Brucella.” Mol. Microbiol., 1998, 27(1): 99-106
(10) Lezhava A., Mizukami, T., Kajitani, T., Kameoka, D., Redenbach, M., Shinkawa, H., Nimi, O. and Kinashi, H. “Physical Map of the Linear Chromosome of Stremptomyces griseus.” Journal of Bacteriology, 1995, 177(22): 6492-6498
(11) Suwanto, A. and Kaplan, S. “Chromosome Transfer in Rhodobacter sphaeroides: Hfr Formation and Genetic Evidence for Two Unique Circular Chromosomes.” Journal of Bacteriology, 1992, 174(4): 1135-1145
(12) Suwanto, A. and Kaplan, S. “Physical and Genetic Mapping of the Rhodobacter sphaeroides 2.4.1 Genome: Presence of Two Unique Circular Chromosomes.” Journal of Bacteriology, 1989, 171(11): 5850-5859
(13) Ussery, D. “Bacteriophage Genetics.” 2000, http://www.cbs.dtu.dk/staff/dave/roanoke /genetics980311.html
(14) Volff, J.N. and Altenbuchner, J. “A New Beginning with New Ends: Linearisation of Circular Chromosomes during Bacterial Evolution.” FEMS Microbiology Letters, 2000, 186: 143-150
(15) Winterstein, C. and Ludwig, B. “Genes Coding for Respiratory Complexes Map on All Three Chromosomes of the Paracoccus denitrificans Genome.” Arch. Microbiol., 1998, 169(4): 275-281
(16) Yamaichi, Y., Iida, T., Park, K.S., Yamamoto, K. and Honda, T. “Physical and Genetic Map of the Genome of Vibrio parahaemolyticus: Presence of Two Chromosomes in Vibrio Species.” Mol Microbiol., 1999, 31(5): 1513-1521