Aichi virus: Difference between revisions
No edit summary |
|||
| Line 1: | Line 1: | ||
{{Viral Biorealm Family}} | {{Viral Biorealm Family}} | ||
[[Image: | [[Image:aichi.jpg|thumb|200px|right|<i>Apis mellifera</i>, a common host of Kashmir bee virus. [http://www.ars.usda.gov/IS/pr/2007/071119.htm].]] | ||
<br><br><br> | <br><br><br> | ||
Revision as of 02:17, 2 May 2013
A Viral Biorealm page on the family Aichi virus
Baltimore Classification
The Aichi Virus is a (+) ssRNA .
Higher order categories
Viruses; ssRNA viruses; ssRNA positive-strand viruses, no DNA stage; Picornavirales; Picornaviridae; Kobuvirus; Aichi Virus
Description and Significance
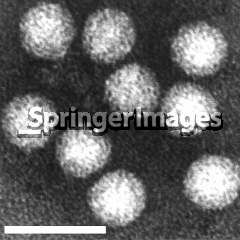
The Aichi Virus is a member of the Kobuvirus genus in the Picornaviridae family. It is a positive sense, single-stranded RNA virus. The virus Aichi Virus appears to morphologically resemble astroviruses when examined by electron microscopy .
Genome Structure
The Aichi Virus is a RNA virus with a genome of 8,280 nt and a poly (A) tail. The virus also can be divided into 2 genotypes, A and B, with around 90% sequence homology .
Virion Structure
The stem-loop at the 5' end of the Aichi Virus genome is an element involved in both viral RNA replication and production of infectious virus particles .
Reproductive Cycle in a Host Cell
Reproduction occurs when there is successful attachment of the virus to a host’s receptors. Host receptors help with endocytosis of the virus into the cell. The capsid of the of the virus changes and is thought to open a pore within the host's endosomal membrane and the viral genomic RNA infiltrates the host cell cytoplasm. A protein, VPg, is taken out from the viral RNA and translated into a processed polyprotein. Replication happens in "viral factories" from the membrane vesicles resulting from the ER. A double stranded RNA genome is made from the genomic ssRNA (+). The dsRNA genome is transcribed/replicated thereby providing vial mRNAs/new ssRNA (+) genomes. It is thought that new genomic RNA is put into assembled procapsids. The cell lyses and the viruses are released . The virus’s L protein is needed for viral RNA replication. There has been ongoing research about the significance of the L protein, which is important for viral RNA replication and excapsidation within virus replication. This is a unique aspect of the Aichi Virus compared to other Picornoviruses .
Viral Ecology & Pathology
Not much is known about the epidemiology of the virus. The Aichi Virus was first found in 1989 as the cause of oyster-associated non-bacterial gastroenteritis in man . It has also been isolated from Japanese travelers in Southeast Asia and has been found in fecal specimens of children with diarrhea in several countries like Brazil, France, Germany, and Tunisia . Not much is known about the virulence factors.
References
Example: J Bacteriol. 2005 May; 187(9): 3188–3200. doi: 10.1128/JB.187.9.3188-3200.2005
Aichi Virus human/hun298/2000/hun. (n.d.). Retrieved from http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=571505&lvl=3&lin=f&keep=1&srchmode=1&unlock
Sasaki, J. (2001). Construction of an infectious cdna clone of Aichi Virus (a new member of the family picornaviridae) and mutational analysis of a stem-loop structure at the 5' end of the genome. J Virol,75(17), 8021-30. , doi: 10.1128/JVI.75.17.8021-8030.2001
Viralzone:kobuvirus. (n.d.). Retrieved from: http://viralzone.expasy.org/viralzone/all_by_species/652.html
Yang S, Zhang W, Shen Q, Yang Z, Zhu J, Cui L, et al. Aichi Virus strains in children with gastroenteritis, China [letter]. Emerg Infect Dis [serial on the Internet]. 2009 Oct [12 Feb 2013].
Yamashita, T. (1998). Complete nucleotide sequence and genetic organization of Aichi Virus, a distinct member of the picornaviridae associated with acute gastroenteritis in humans. JVirol, 72(10), doi: http://jvi.asm.org/