Alistipes finegoldii: Difference between revisions
| Line 69: | Line 69: | ||
[11][https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/ijs.0.64318-0 ''Alistipes onderdonkii sp. nov. and Alistipes shahii sp. nov., of human origin''] | [11][https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/ijs.0.64318-0 ''Alistipes onderdonkii sp. nov. and Alistipes shahii sp. nov., of human origin''] | ||
[12] [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2828066/ ''Alistipes finegoldii in Blood Cultures from Colon Cancer Patients''] | [12][https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2828066/ ''Alistipes finegoldii in Blood Cultures from Colon Cancer Patients''] | ||
[13][https://pubmed.ncbi.nlm.nih.gov/21742925/ ''Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials''] | |||
[14][https://pubmed.ncbi.nlm.nih.gov/30574320/ ''Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease''] | |||
[15][https://pubmed.ncbi.nlm.nih.gov/25758642/ ''Gut microbiome development along the colorectal adenoma-carcinoma sequence''] | |||
[16][https://pubmed.ncbi.nlm.nih.gov/30951169/ ''The effect of diet on hypertensive pathology: is there a link via gut microbiota-driven immunometabolism?''] | |||
[17][https://pubmed.ncbi.nlm.nih.gov/29507058/ ''Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure''] | |||
==Author== | ==Author== | ||
Revision as of 02:12, 23 April 2024
Classification
Higher order taxa
Domain: Bacteria
Phylum: Bacteroidota
Class: Bacteroidia
Order: Bacteroidales
Family: Rikenellaceae [1]
Species
Alistipes finegoldii NCBI: Taxonomy
Description and Significance
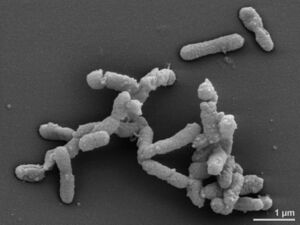
Alistipes finegoldii is a commensal anaerobe that is gram-negative, rod-shaped, and non spore forming bacteria [2] that are found primarily in the gastrointestinal tract [2]. Alistipes finegoldii was first discovered in 2003 by Sydney M. Finegold, a researcher of anaerobic bacteriology [2]. The species was subsequently named after him. Alistipes is a relatively new genus of bacteria and the first species, Alistipes finegoldii, was found in tissue samples of children with appendicitis [4]. So far, the Alistipes genus contains 13 species, all of which besides A.obesi are non-motile [2]. The discovery of Alistipes finegoldii bacteria has become essential in clinical research, as it has been shown to have both possible protective effects against diseases and pathogenetic dysbiotic effects. Studying Alistipes finegoldii can provide more insight on the relationship of bacteria-host symbiosis in the gut and other areas of the body as well. Furthermore, its continuous study will contribute to how we understand Alistipes finegoldii and its relationship to human health [2]. Because Alistipes finegoldii are bile-resistant, they are more likely to be present in the terminal ileum where most bile reabsorption occurs and where chronic inflammatory bowel conditions are more likely to occur [2]. Because of how recent the discovery of Alistipes is, more studies will need to be done to determine if bile-resistance is the reason Alistipes is abundant within the GI tract or in diseases that are characterized by alterations in bile production [2].
Genome Structure
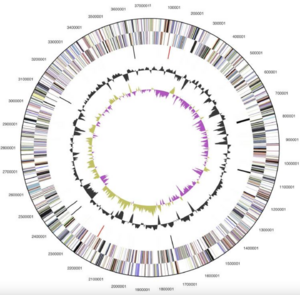
Alistipes finegoldii (type-strain AHN2437T [3,12]) was the first species of the Alistipes genus whose genome was completely sequenced. This bacteria consists of 1 circular chromosome and is 3,734,239 bp in length [3]. Of the 3,302 total genes, 3,234 of them are protein coding and 68 are RNA genes [3]. Additionally, 121 pseudogenes were found [3]. Alistipes finegoldii’s coding region is 86.89% of its genome, 56.5% of which is its G+C content [1,3].
The Alistipes finegoldii genome was selected for sequencing as the first species in the Alistipes genus classified in 2003 [3,11]. 16S rDNA sequencing studies showed that Alistipes finegoldii clustered with Alistipes putredinis in the Bacteroidetes group [12]. In a 16S rRNA gene sequence of Alistipes finegoldii, it was found that Alistipes shahii and Alistipes onderdonkii yielded the highest scores, with a divergence of less than 3% and a match with Alistipes finegoldii at approximately 97% [3,11]. These genomic divergences tell us that Alistipes finegoldii has a distinct role and carries out different functions as compared to other species of the Alistipes genus.
Cell Structure, Metabolism and Life Cycle
Alistipes finegoldii is a chemoorganotroph, meaning they get their energy from the oxidation of organic compounds. This also means they obtain hydrogen and/or electrons from organic compounds such as fats, sugars, and proteins. It is speculated that A. finegoldii has a fermentative metabolism [1], but it has been hard to determine due to its difficult growth in vitro in agar and liquid media [3]. This bacteria is singularly occurring, but occasionally A. finegoldii has been observed to have longer filaments [3]. It is a strictly anaerobic mesophile with an incubation period of 1-2 days at 37ºC [1,3]. Additionally, A. finegoldii is indole-positive and has shown to be gamma and/or weakly beta hemolytic [3]. One novel trait of this bacteria is that it is bile-resistant, meaning it has a resistance to or is able to cope with bile salts. This resistance is the efflux of bile salts from bacterial cytoplasm directly out of the cell wall, and is a characteristic property of probiotic bacteria. By looking at the lipid biosynthetic pathways of Alistipes finegoldii, it was found that this species only produces saturated fatty acids, the most abundant of which are phosphatidylethanolamine (PE) and sulfonolipid (SL) [8]. Furthermore, the utilization of these fatty acid chains along with their correlating synthases allows Alistipes finegoldii to assemble its membrane lipids in the gut environment [8].
Ecology and Pathogenesis
Habitat; symbiosis; biogeochemical significance; contributions to environment.
If relevant, how does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
(talk about how it can be become pathogenic in some cases or protective, it metabolism, insert pic from one of the main studies, etc.
Alistipes finegoldii is found ubiquitously in the gastrointestinal tract...... There is contrasting evidence regarding Alistipes pathogenicity. Recent studies show that there are links between Alistipes and the protection against colitis, liver fibrosis (cirrhosis), and cardiovascular disease. On the other hand, different studies have found that Alistipes is the pathogen responsible for colorectal cancer and is linked to mental health issues such as depression. (explain each case) T
ecology part describe how its prepped in the lab and the env it grows in, how it mimics this env, ecological advantage?
References
[5]Gut microbial carbohydrate metabolism contributes to insulin resistance
[7]Sulfonolipids as novel metabolite markers of Alistipes and Odoribacter affected by high-fat diets
[11]Alistipes onderdonkii sp. nov. and Alistipes shahii sp. nov., of human origin
[12]Alistipes finegoldii in Blood Cultures from Colon Cancer Patients
[15]Gut microbiome development along the colorectal adenoma-carcinoma sequence
Author
Page authored by Virginia Powell & Max Plodzik, students of Prof. Jay Lennon at Indiana University.