Altiarchaeales: Difference between revisions
Chadalavadas (talk | contribs) |
|||
| Line 4: | Line 4: | ||
<br>By Benjamin A. Canniff<br> | <br>By Benjamin A. Canniff<br> | ||
<br>The topic of my project is the newly discovered organism dubbed Altiarchaeales. | <br>The topic of my project is the newly discovered organism dubbed Altiarchaeales. The objective of this project is to provide a in depth guide to understanding these new organisms, and how they fit into out environment.<br> | ||
==Higher Order Taxa== | ==Higher Order Taxa== | ||
Revision as of 17:33, 28 April 2017
Introduction
By Benjamin A. Canniff
The topic of my project is the newly discovered organism dubbed Altiarchaeales. The objective of this project is to provide a in depth guide to understanding these new organisms, and how they fit into out environment.
Higher Order Taxa
Kingdom:Proteoarchaeota
- Domain: Archaea
- Phylum: Euryarchaeota
- Class: Unknown
- Order: Altiarchaeales
- Family: Altiarchaeaceae
- Order: Altiarchaeales
- Class: Unknown
- Phylum: Euryarchaeota
Species
- Genus: Altiarchaeum
- Species: Archaea
Description and Significance
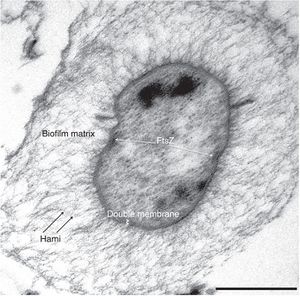
Altiarchaeles are an order of uncultured Archaea that is a recently proposed order. The Altiarchaeles’ lineage is made up of genetically diverse and globally wide spread microorganisms that can be found in anoxic subsurface environments, and are one of the few archaea that have a double membrane. The entire cell wall consists of an outer membrane separated by a periplasmic space from the cytoplasmic membrane. In depth genome analysis suggests that Altiarchaeles are autotrophic, utilizing a modified version of the archaeal reductive acetyl-CoA pathway, otherwise known as the Wood-Ljungdahl pathway for metabolism (Figure 2)[4].
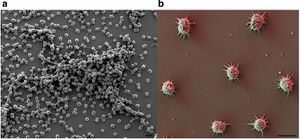
The Archaea was first discovered over 15 years ago in a swamp located in Germany, where scientists noticed a so-called string of pearls microbe community floating on the surface of the water. They took samples of these colonies and found that the inner part of the string of pearls was made up of a new microorganism, now known as Canididatus Altiarchaeum hamiconexum. The outer part of the pearls turned out to be a filamentous sulfur-oxidizing microorganism called Thiothrix. Another group of Ca. A. hamiconexum was discovered in a drilled 36.5 m deep hole that had sulfidic ground water coming from it. However unlike before, where the Ca. A. hamiconexum was in a string of pearls colony formation, they found a biofilm composed almost purely of just Ca. A. hamiconexum. At that point in time an archaea that had the ability to form a highly-pure biofilm in the natural environment had not been discovered yet, making Ca. A. hamiconexum the first of its kind. The size and purity of the biofilm found indicated that the Ca. A. hamiconexum not only were metabolically active but also had a high efficiency at reproducing. The biofilm also seems to be able to out compete or suppress other organisms by some unknown means, leading the high purity of the biofilm (Figure 3)[4].
By some unknown mechanism, parts of the biofilm are released and transported to the surface by the water’s current, making this the first archaeal organism that undergoes a change in its lifestyle while remaining in its natural habitat. When these parts reach the surface, they form a companionship with the sulfur-oxidizing Thiothrix and form the string of pearls colony. Scientist also found that the lower the oxygen concentration in the body of water, the greater the abundance of Ca. A. hamiconexum present in the ecosystem. The unique characteristic of the Ca. A. hamiconexum is its appendage called the hami that acts like a grappling hook, anchoring the string of pearls colony to a solid object. This anchor allows the colony to stay stationary on top of the water even if there is a current. These hami appendages are unique to Ca. A. hamiconexum and have no analog in other microbes. Once anchored above the water, the archaea proliferate for a few days before the string of pearls comes apart and the microbes disperse into the water. Even though these hami have some similarities to flagella and other pili, altiarchaeales are a non-motile archaea. Unfortunately to this day the microorganism still hasn’t been successfully cultured, instead relying on cultivation from mass string of pearls communities created in nature through the use of polyethylene nets (Probst, 2014). Scientists have not been able to recreate in lab the specific environment conditions that are needed for Ca. A. hamiconexum to grow. These conditions being a very high flow rate of water rushing past it, around 5400 L/minute, which makes the estimation of nutrient availability and waste product concentrations to be extremely difficult to estimate. However the discovery of Altiarchaeales has revealed that the subsurface can harbor lineages of microbial life that are still vastly unexplored. Hopefully through studying the Altiarchaeales’ genomic and phylogeny data scientists can piece together lineages and discover completely new organism living deep below the surface [4].
Structure of Biofilm and Hami Appendages
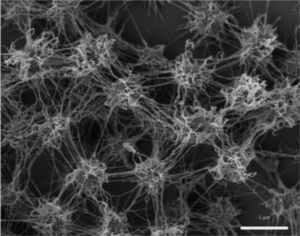
The biofilm is formed by the coccoid cells with a diameter of approximately 0.6 μm. The coccoid cells make the biofilm porous but also rigid and well structured, where the cells are arranged in three dimensional patterns 2 μm apart from each other. A protein and carbohydrate-rich matrix with a thickness of 600 nm also encases the cells (Figure 4) [4]. Gluconeogenesis products like glucose and fructose are used for synthesis of a variety of activated forms of sugars, these sugars serve as the building blocks for the synthesis of the biofilm polymers. The polymers that make up the biofilm are comprised of mostly carbohydrates. The well-equipped sugar biosynthesis machinery that all Altiarchaeales possess is an essential adaptation to allowing the organisms to dominate the subsurface biotopes that they reside in. The biofilm dominates the subsurface biotopes by acting like a filter that blocks the unwanted organisms from entering their specific area. It has also been hypothesized that the biofilm could possible serve a role in electron transfer in the cells. This dominance over other organism that lives in those subsurface anaerobic environments is not observed in other archaeal lineages. As such it can be speculated that this archaeal dominance could be commonplace in these deep subsurface anaerobic environments [2].
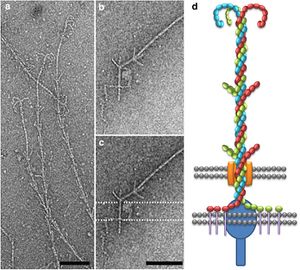
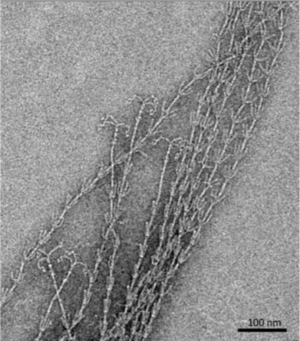
The hami, a filamentous cell surface appendage, facilitates the connections between the cells in the biofilm and anchors the cells to other objects in their environment through van der Waals forces. The “string” in the string of pearls colonies are the hami holding together the Altiarchaeales. The entire structure of the hami resembles a barbed wire running approximately 5 μm in length with a barbed grappling hook at the end of the structure. The hami are anchored into the cell past its double membrane and into the cytoplasm. The grappling hooks at the end of the hami are the smallest biologically-formed hooks ever discovered, and are somehow made thicker than the rest of the structure, making them the strongest part of the hami (Figure 5) [3].The main shaft of the hami is actually composed of helical proteins wound together around each other, and wherever a barb appears along the length of the hami indicates the start of a helical twist. The prickles that show up along the hami assume an angle of 45-60 degrees and point towards the grappling hook end of the hami. Hami from multiple different cells were observed and each time the hami always showed the same basic structure, dimensions, and proportions. The hami is also composed of only 1 major protein species, showing no similarities to known microbial structures. Furthermore the proteins do share similarities via its protein sequence and structure to S-layer proteins, proteins located along the surface of the cell, of archaea. These similarities also raise the question on whether the hami evolved from these S-layer proteins or vice versa (Figure 6) [3].
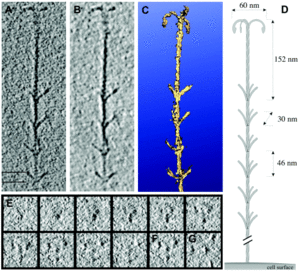
Hypotheses of other uses of the hami, beside attachment, have been proposed, one being when attached to another cell it establishes a connection, starting an intra-cellular signal cascade. This communication hypotheses would make the most sense when the string of pearls colonies are formed, for the first connection between the Altiarchaeales and bacteria are the hami, making it likely that some form of communication occurs through the hami and the two cells that are attached to it [4].Even though the hami are composed of extremely small proteins wrapped around each other, they are actually incredibly strong. They can remain stable at temperatures between 0-70 °C and PH values from 0.5-11.5, becoming unstable at 80 °C and PH of 12.5. However, just like any other protein based structure, the hami are very sensitive to enzymatic treatments with protease. Through adhesion tests scientists found that the hami could bind to a variety of different surfaces with different chemical properties. Examples being polylysine, polyglutamate, gelatin, bovin serum albumin, laminin, fibronectine, and bind-silane [3]. Remarkably the hami adhered to all the surfaces that it was tested against, showing no weakness to any particular one. Hami are also extremely different from other prokaryotic appendages such as pili and flagella. Both pili and flagella have a far less complicated architecture than the hami, instead of multiple different strands combined in a helical formation the pili and flagella are made up of form tubes and helical fibers. Furthermore pili and flagella have a hollow central channel used for signal transferring whereas the hami do not. Scientists are still baffled at how the barb wire like structure is synthesized, they believe the hami to grow via cell addition at the base, but they are still very much in the unknown about how those barbs and hooks take form. They hypothesize that, because of the hami’s complexity, a multicomponent system is the only thing that can control and coordinate the building of something so complicated. Because of their extremely unique and sturdy structure the hami’s nanostructured surfaces could lead into new discoveries and advances in the field of Nano biotechnology [3].
Metabolism
Ca. A. hamiconexum are an autotrophic organism that uses a modified reductive acetyl-CoA pathway to turn carbon dioxide into organic compounds. This reductive acetyl-CoA pathway is also known as the Wood-Ljundhal pathway, and can only be accomplished under anaerobic conditions, the exact type of locations that Ca. A. hamiconexum have been found. Usually the Wood-Ljundhal pathway is coupled with enzymes and proteins that are involved in methanogenesis as well, however Ca. A. hamiconexum has been found to contain none of the enzymes essential for methanogenesis to occur. The archaeal version of the Wood-Ljundhal pathway uses tetrahydromethanoplerin, a coenzyme acting as the C1 carrier in methanogenesis. The modified version of this pathway comes from replacing dehydrogenase and reductase of methylene-tetrahydrofolate with analogous enzymes working with NAD(P)H (Figure 7) [4].

The generated acetyl-CoA ultimately undergoes gluconeogenesis, leading to the production of multiple types of sugars. The exact electron donor and acceptors are still relatively unknown. It has been hypothesized that carbon monoxide and hydrogen gas, both prevalent in most groundwater ecosystems, could act as electron donors for Ca. A. hamiconexum. Ferrous iron, found abundantly in the Altiarchaeales environment, could also be a potential electron donor. Another possibility being that the electrons could be passed on to other microorganisms such as the sulfate reducing bacteria that seem to have a companionship relationship established between the two. This lack of knowledge, pertaining to the unknown electron donors and acceptors, is another key reason why Altiarchaeales cannot be cultured yet, for most archaea specializes in surviving at the knife edge of metabolism. As such, without knowing the exact amount of whatever compound it uses for electron donors and acceptors the organism cannot sustain itself [2]. Other hypothesizes that involve the oxidation of ferredoxin or the use of the hami structure in the generation of a proton gradient, however nothing has been proven as of yet, and the majority of the organism’s actual metabolism process is still unknown. The only concrete fact that is known about the metabolism process is that the oxidation of ferredoxin appears to be central in the establishment of a proton gradient for energy conservation. This lack of knowledge in how exactly they metabolize energy is one of the major issues why Altiarchaeales have yet to be cultured, for they cannot design culture conditions that mimic its original habitat without knowing what essential nutrients and compounds it needs to survive [3].
Phylogeny
Gathering information on the lineage of the Ca. A. hamiconexum has been made difficult by only having genomic data from two representatives of the species, one from the marshland in Germany, and the other from an underground water ecosystem in the US. Because of these different environments that the samples were found in, scientists identified two major sequence clades within Altiarchaeales, Alti-1 and Alti-2. Alti-1 represents the Altiarchaeales that are found in spring environments, whereas Alti-2 sequences were derived from a variety of different anoxic environments. By investigating the placement of multiple protein markers, 56 concatenated ribosomal proteins from 142 archaeal genomes, along with using a phylogenomics approach based on concatenated ribosomal protein sequences and state of the art treeing methods it has been discovered that Ca. A. hamiconexum form a robust monophyletic cluster residing on a long branch, revealing a sister relationship to the species methanococcales (Figure 8) [1].
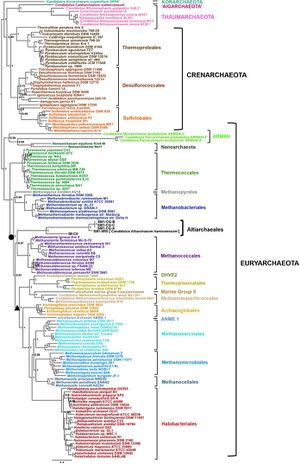
This relationship with methanococcales is the only close relationship that Ca. A. hamiconexum has with another organism. Furthermore Ca. A. hamiconexum has been found to not have any close relationship to an archaeal lineage. Through investigation of Ca. A. hamiconexum’s genes, specifically the 16s rRNA gene it was discovered hat around 25.8 % of their genes were potentially acquired through horizontal gene transfer between bacteria. Other, more distant, relatives of the organism were found in hot springs, ponds, and deep sea ecosystem. Ultimately the phylogenetic analyses indicate that Ca. A. hamiconexum are representatives of a new Euryarchaeal order-level lineage of anaerobic, aquatic subsurface archaea [1].
Wood-Ljundhal Pathway
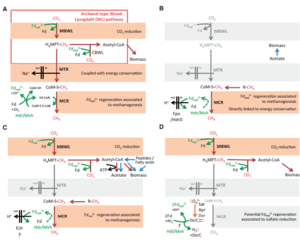
The Wood-Ljundhal pathway is an extremely important pathway for metabolism for energy generation and carbon fixation. This form of metabolism is used by archaea and bacteria, and is mostly seen coupled with methanogenesis. The archaeal and bacteria that perform the wood-Ljundhal pathway involve different C1-carriers, cofactors, electron transporters, and enzymes. In bacteria the end product of this process is acetate, whereas archaea produce methane, leading to the idea that methanogenesis and Wood-Ljundhal are linked. In most cases carbon dioxide and hydrogen gas are the starting materials of this pathway, where carbon dioxide is reduces to a methyl group using ferredoxin. From the creation of the methyl the reaction can go either of two ways, the methyl group could be transferred to tetrahydromethanopterin and then reduced again forming acetyl-CoA, leading to biomass production. The other option being that the methyl group is transferred to the coenzyme M by the MTR (methyltransferase complex). This transfer displaces the sodium gradient outside the cytoplasmic membrane and as a result ATP synthase is activated, generating ATP that becomes stored energy. The MCR (methyl-coenzyme M reductase complex) then catalyzes the formation of methane and heterodisulfide, releasing then as waste products (Figure 9) [5].
Genomics
A genomic study of seven different samples of Altiarchaeales was done and from this study they discovered only 57 genes were homologous to all seven of the samples, but about 573 genes were shared between most of the samples. The seven samples were part of either the Alti-1 or Alti-2 clade system. The scientists that performed this experiment referred to these common 573 genes as the core Altiarchaeales genes. Even though many of those 573 genes are conserved universally throughout archaea there were a few specific to the Altiarchaeales (Bird, 2016). These genes contain the information for processes such as the TCA cycle, transcription and translation, glycolysis, pyruvate synthase, and acetyl-CoA synthase. However they also found genes that encoded for dehydrogenase ferredoxin, the key process for biosynthesis of amino acids, sugars, lipids, and cobalamins. Furthermore genes that are involved in the Wood-Ljundhal processes such as the carbon dioxide fixation 5,10-methylenetetrahydrofolate reductase and the NADPH dependent methylenetetrahydromethanopterin dehydrogenase. All of these Wood-Ljundhal genes are what the scientists expected to see, considering that they are also present in most archaea (Figure 10)[1].
However the gene that encodes for NADPH dependent methylenetetrahydromethanopterin dehydrogenase is not found in archaea and is more similar to genes found in bacteria. They also found that all Alti-1 and Alti-2 genomes have the same set of replication genes, along with the same replication machinery. Furthermore through their study of the replication genome they discovered a mutation, which was observed in six out of the seven samples, in the replication machinery phenylalanine tRNA beta subunit. This specific beta subunit dealt with histidine synthase in the cells. Through tracing the path of this mutation they believe the site of this mutation to be a fission event. This fission event is extremely similar to the split in the Nanoarchaeota tRNA-binding domain site, again affecting two different tRNAs, preventing them from fulfilling their task. This similarity between Nanoarchaeota and Altiarchaeales led the scientists to believe that these fission events represented two different evolutionary events. This evolutionary event is most likely due to a split between two protein domains that occurred in an ancestor of the two organisms. Moreover this discovered mutation, evolutionary split, and common ancestor helps define the placement of the Altiarchaeales. Genes corresponding to biofilm production were also found, however Alti-1 organisms, the Altiarchaeales found in the German swamp, contained more of these biofilm producing genes than the Alti-2 organisms. This difference is most likely due to the Atiarchaeales specific location, where biofilm production would be more beneficial to the organisms living in those swamp like environments. Where Alti-2 lacks in biofilm production it makes up with containing many sulfur related genes, which are probably for sulfur assimilation. These sulfur related genes make sense considering the Alti-2 organisms residing deep in underground wells where sulfur is high in concentration. One set of genes that were seen throughout both the Alti-1 and Alti-2 organisms are the genes associated with the production of hami. Other than in Altiarchaeales these gene sequences have no homologues in public databases and are considered to be only found in Altiarchaeales [1].
References
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2017, Kenyon College.
