Bacillus cereus
A Microbial Biorealm page on the genus Bacillus cereus
Classification
===Higher order taxa===
Domain: Bacteria
Phylum: Firmicutes
Class: Bacilli
Order: Bacillales
Family: Bacillaceae
Genus: Bacillus
Species Group: Bacillus cereus group
Species
|
NCBI: Taxonomy |
Bacillus cereus
Description and significance
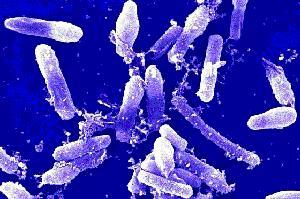
Bacillus cereus is a large, 1 x 3-4 µm, gram-positive, rod-shaped, endospore forming, facultative aerobic bacterium [2]. It was first successfully isolated in 1969 from a case of fatal pneumonia in a male patient and was cultured from the blood and pleural fluid [5]. 16s rRNA comparison reveals Bacillus cereus to be most related to Bacillus anthracis, the cause of anthrax, and Bacillus thuringiensis, an insect pathogen used as pesticide [3]. Although they have similar characteristics they are distinguishable with B. cereus being most motile, B. thuringiensis produce crystal toxins, and B. anthracis is nonhemolytic [4].
B. cereus is a mesophile, growing optimally at temperatures between 20 and 40°C, and is capable of adapting to a wide range of environmental conditions. It is distributed widely in nature where it is most commonly found in the soil as a saprophytic organism. B. cereus is also an inhabitant of the microflora of insects as a gut commensal, deriving nutrients from its host, and is found in the rhizosphere of some plants [2]. B. cereus spores and cells are released back into the soil where they germinate upon defecation or death of their host until they are ingested by another host.
As a soil bacterium, B. cereus can spread easily to many types of foods such as meat, eggs, dairy products, and plants, and is commonly known for causing 25 % of food-borne intoxications due to is production of emetic toxin and enterotoxins [4]. Food poisoning occurs when food is left without refrigeration for several hours before it is served. Surviving spores of contaminated foods after heat treatment grows well and is the source of the food poisoning [4].
In addition, Bacillus cereus is an opportunistic human pathogen and is occasionally associated with infections, causing periodontal diseases and other more serious infections [5]. Immunocompromised individuals are susceptible to bacteremia, endocarditis, meningitis, pneumonia, and endophthalmitis [6]. Its potential to cause systemic infections are of current bio-medical and public health concerns. Thus, the genome sequence of Bacillus cereus is significant in establishing genetic background information for further investigations and to promote public health preparedness. Sequencing its genome is vital to expand the understanding of its pathogenicity, and for the development of anti-microbial drugs. Additionally, since Bacillus cereus strains are so genetically closely related to B. anthracis, used in biological warfare, and can display anthrax-like virulence traits, genomic comparisons between the two species is important in order to study the virulence of B. anthracis to produce vaccines and prevent its usage as a bioweapon.
Genome structure
1.1 Genome
B. cereus has a circular chromosome measuring 5,411,809 nt in length and was completely sequenced using the shotgun method [7]. The genome structure of Bacillus cereus consists of 5481 genes, 5234 protein coding, 147 structural RNAs, and 5, 366 RNA operons [1]. An interesting gene cluster found within the genome of Bacillus cereus is a gene that encodes for arginine deiminase metabolic pathway. The presence of this cluster is predicted to have a role in its survival, enabling B. cereus to be resistant to acidic conditions in a similar manner Streptococcus pyogenes does [8]. Additionally, B. cereus has a nine-gene urease gene cluster that encodes for proteins, blasticidin S deaminase , and S layer protein [8]. The urease enzyme has a role in increasing vigor of B.cereus in acidic conditions, which is similar to the Helicobacter pylori urease that is required for colonization of the human stomach [8].
Genes present within the chromosomes associated with B. cereus virulence include non-haemolytic enterotoxins, channel-forming type III hemolysins, phospholipase C, a perfringolysin O (listeriolysin O), and extracellular proteases [6]. All three proteins of the Haemolysin BL enterotoxins associated with food poisoning are transcribed from one operon (hbl), which is an RNA transcript of 5.5 kb. These genes along with other genes encoding for metabolic enzymes, proteins involved in motility and chemotaxis, proteins involved in sporulation, and cellular transporters are all regulated by the plcR gene [9]. In addition to its role in the regulation of transcription of these genes for survival, the plcR gene is responsible for the full virulence of B. cereus. Therefore, it is
often the target of antimicrobial agents. Another gene found on its chromosome is the gerA gene which is essential to the sporulation of B. cereus when nutrients are depleting, and is responsible for spore germination in response to l-alanine and ribosides [9].
B. cereus have fewer genes for carbohydrate catabolism and more genes for amino acid metabolism which suggests that they are susceptible to environments that are rich in protein, having only twelve carbohydrate polymer degradation genes [9]. Furthermore, proteins, peptides, and amino acids appear to be preferred nutrient sources for B. cereus with 18-23 genes that encode peptide and amino acid ABC transporter-ATP binding proteins [9].
1.2 Plasmids
B. cereus has a diverse range of plasmids that vary in size from 5 to 500 kb, and is known to have more than one plasmid with only a few plasmids that are associated with pathogenesis. B. cereus ATCC has a large pXO1 plasmid that is also found in B. anthracis. It is not virulent because it lacks the portion that encodes the lethal toxin and regulatory proteins [7]. B. cereus G9241 has a plasmid that is 99.6% identical to pXO1 plasmid from B. anthracis, but does not have the pXO2 plasmid. It also has an additional plasmid that encodes for a capsule biosynthesis operon [7]. B. cereus ZK, a pathogenic strain of bacteria have five plasmids and is the largest amount of plasmids found in a strain of B. cereus yet. A large number of transposase genes are found in the two large plasmids of this strain assisting in gene exchange between plasmids and chromosome [7]. The three small plasmids are believed to function in the identification of replication and mobilization [7].
Cell structure and metabolism
Describe any interesting features and/or cell structures; how it gains energy; what important molecules it produces.
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
Enter summaries of the most recent research here--at least three required
References
[1] "Bacillus cereus." NCBI website. Accessed on August 18, 2007
[2] Vilain, S., Luo, Y., Hildreth, M., and Brozel, V. “Analysis of the Life Cycle of the Soil Saprophyte Bacillus cereus in Liquid Soil Extract and in Soil.” Applied Environmental Microbiology. 2006. Volume 72(7). p. 4970–4977.
[3] DelVecchio, V., Connolly, J., Alefantis, T., Walz, A., Quan, M., Patra, G., Ashton, J., Whittington, J., Chafin, R., Liang, X., Grewal, P., Khan, A., and Mujer C. “Proteomic Profiling and Identification of Immunodominant Spore Antigens of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis.” Applied Environmental Microbiology. 2006. Volume 72(9). p. 6355–6363.
[4] “Bacillus cereus.” United States Food and Drug Administration, Center for food safety and applied nutrition (FDA). Accessed August 18, 2007.
[5] Hoffmaster, A., Hill, K., Gee, J., Marston, C., De, B., Popovic, T., Sue, D., Wilkins, P., Avashia, S., Drumgoole, R., Helma, C., Ticknor, L., Okinaka, R., and Jackson, J. “Characterization of Bacillus cereus Isolates Associated with Fatal Pneumonias: Strains Are Closely Related to Bacillus anthracis and Harbor B. anthracis Virulence.” Journal of Clinical Microbiology. 2006. Volume 44(9). p. 3352-3360.
[6] Wijnands, L., Dufrenne, J., Zwietering, M. H., and Leusden, F. “Spores from mesophilic Bacillus cereus strains germinate better and grow faster in simulated gastro-intestinal conditions than spores from psychrotrophic strains.” International Journal of Food Microbiology. 2006. Volume 112. Issue 2. p. 120-128.
[7] Rasko, D., Altherr, M., Han, C., and Ravel, J. “Genomics of the Bacillus cereus group of organisms.” FEMS Microbiology Reviews. 2005. Volume 29(2). p.303-329.
[8] Rasko, D., Ravel, J., Okstad, O. A., Helgason, E., Cer, R., Jiang, L., Shores, K. A., Fouts, D., Tourasse, N., Angiuoli, S., Kolonay, J., Nelson, W., Kolsto, A, Fraser, C., and Read, T. D. “The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1” Nucleic Acids Research. 2004. Volume 32(3). p. 977–988.
[9] Han, C., Xie, g., Challacombe, J., Altherr, M., Bhotika, S., Bruce, D., Campbell, D., Campbell, M., Chen, J., Chertkov, O., Cleland, C., Dimitrijevic, M., Doggett, N., Fawcett, J., Glavina, T., Goodwin, L., Hill, K., Hitchcock, P., Jackson, P., Keim, P., Kewalramani, A., Longmire, J., Lucas, S., Malfatti, S., McMurry, K., Meincke, L. J., Misra, M., Moseman, B. L., Mundt, M., Munk, C., Okinaka, R. T., Parson-Quintana, B., Reilly, L. P., Richardson, P., Robinson, D. L., Rubin, E., Saunders, E., Tapia, R., Tesmer, J. G., Thayer, N., Thompson, L. S., Tice, H., Ticknor, L., Wills, P., Brettin, T., and Gilna, P. “Pathogenomic Sequence Analysis of Bacillus cereus and Bacillus thuringiensis Isolates Closely Related to Bacillus anthracis.” Journal of Bacteriology. 2006. Volume 188(900). p. 3382–3390.
[10] Ticknor, O., Kolsto, A., Hill, K., Keim. P., Laker, M., Tonks, M., and Jackson, P. “Fluorescent Amplified Fragment Length Polymorphism Analysis of Norwegian Bacillus cereus and Bacillus thuringiensis Soil Isolates.” Applied Environmental Microbiology. 2001. Volume 67(10). p. 4863–4873.
[11] Amano, K., Hazama, S., Akarari, Y., Ito, E. “Isolation and Characterization of Structural Components of Bacillus cereus AHU 1356 Cell Walls.” European Journal of Biochemistry. (1977). Volume 75 (2). p. 513–522.
[12] Severin, A., Tabei, K., Tomasz, A. “The structure of the cell wall peptidoglycan of Bacillus cereus RSVF1, a strain closely related to Bacillus anthracis.” Microbial Drug Resistance. 2004. Volume 10(2). p. 77-82.
[13] Mignot, T., Denis, B., Couture-Tosi, E., Kolsto, A., Mock, M., Fouet, A. “Distribution of S-layers on the surface of Bacillus cereus strains: phylogenetic origin and ecological pressure.” Environmental Microbiology. 2001. Volume 3(8). p. 493–501.
[14] Senesi, S., Celandroni, F., Salvetti, S., Beecher, D., Wong, A., and Ghelardi, A. “Swarming motility in Bacillus cereus and characterization of a fliY mutant impaired in swarm cell differentiation.” Microbiology. 2002. Volume 148. p. 1785-1794.
[15] Pol, I., van Arendonk, W., Mastwijk, H., Krommer, J., Smid, E., and Moezelaar R. “Sensitivities of Germinating Spores and Carvacrol-Adapted Vegetative Cells and Spores of Bacillus cereus to Nisin and Pulsed-Electric-Field Treatment.” Applied Environmental Microbiology. 2001. Volume 67(4). p. 1693–1699.
[16] Kutima, P., and Foegeding, P. “Involvement of the spore coat in germination of Bacillus cereus T spores.” Applied Environmental Microbiology. 1987. Volume 53(1). p.47–52.
[17] Duport, C., Zigha, A., Rosenfeld, E., and Schmitt, P. “Control of Enterotoxin Gene Expression in Bacillus cereus F4430/73 Involves the Redox-Sensitive ResDE Signal Transduction System.” Journal of Bacteriology. 2006. Volume 188. p. 6640–6651.
[18] Mols, M., de Been, M., Zwietering, M., Moezelaar, R., Abee, T. “Metabolic capacity of Bacillus cereus strains ATCC 14579 and ATCC 10987 interlinked with comparative genomics.” Environmental Microbiology. 2007. (Online Early Articles).
[19] Silo-Suh, L., Lethbridge, B., Raffel, S J, He, H., Clardy, J., and Handelsman, J. “Biological activities of two fungistatic antibiotics produced by Bacillus cereus UW85.” Applied Environmental Microbiology. 1994. Volume 60(6). p. 2023–2030.
[20] Halverson, L J, and Handelsman J. “Enhancement of soybean nodulation by Bacillus cereus UW85 in the field and in a growth chamber.” Applied Environmental Microbiology. 1991. Volume 57(9). p. 2767–2770.
[21] Peterson, S., Dunn, A., Klimowicz, A., and Handelsman, J. “Peptidoglycan from Bacillus cereus Mediates Commensalism with Rhizosphere Bacteria from the Cytophaga-Flavobacterium Group.” Applied Environmental Microbiology. Volume 72(8). p. 5421–5427.
[22] Margulis, L., Jorgensen, J, Dolan, S., Kolchinsky, R., Rainey, F., and Shyh-Ching. “The Arthromitus stage of Bacillus cereus: Intestinal symbionts of animals.” Proceedings of the National Science Academy U S A. 1998. Volume 3; 95(3). p. 1236–1241.
[23] Jensen, G., Hansen, B., Eilenberg, J., Mahillon, J. “The hidden lifestyles of Bacillus cereus and relatives.” Environmental Microbiology. 2003. Volume 5(8). p. 631–640.
[24 Kotiranta, A., Lounatmaa, K., and Haapasalo, M. “Epidemiology and pathogenesis of Bacillus cereus infections.” Microbes and Infections. 2000. Volume 2, Issue 2. p. 189-198
[25 Granum, P., Lund, T. “Bacillus cereus and its food poisoning toxins.” FEMS Microbiology Letters. 1997. Volume 157 (2). p. 223–228.
[26] Handelsman, J., Jacobson, L., Stabb, E. “Bacillus cereus strain DGA34; United States Patent 5736382.” Official gazette of the United States Patent and Trademark Office. 1998. Accessed August 13, 2007.
[27] Sunaina, V. “Bacterial metabolites from Bacillus cereus B4 responsible for potato plant growth.” Journal of the Indian Potato Association. 2005. Volume 32 (3-4). p. 187-188.
[28] Silo-Suh, L., Lethbridge, B., Raffel, S., He, H., Clardy, J., and Handelsman, J. “Biological activities of two fungistatic antibiotics produced by Bacillus cereus UW85.” Applied Environmental Microbiology. 1994. Volume 60(6) p. 2023–2030.
[29] Wijman, J., Leeuw, P., Moezelaar, R., Zwietering, M., and Abee, T. “Air-Liquid Interface Biofilms of Bacillus cereus: Formation, Sporulation, and Dispersion.” Applied Environmental Microbiology. 2007. Volume 73(5). p. 1481–1488.
[30] Clavel, T., Carlin, F., Dargaignaratz, D., Lairon, D., Nguyen-The, C., Schmitt. P. “Effects of porcine bile on survival of Bacillus cereus vegetative cells and Haemolysin BL enterotoxin production in reconstituted human small intestine media.” Journal of Applied Microbiology. 2007(OnlineEarly Articles).
[31 Harvie, D., Vilchez, D., Steggles, J., Ellar, D. “Bacillus cereus Fur regulates iron metabolism and is required for full virulence.” Microbiology. 2005. Volume 151(Pt 2). p.569-77.
Edited by Jacqueline Nguyen student of Rachel Larsen
