Bacterial phage defense mechanisms with applications: Difference between revisions
(Created page with "{{Uncurated}} =Overview= Bacteria are constantly subjected to bacteriophages and other selfish genetic elements. Bacteriophages are viruses that specifically infect bacteria...") |
No edit summary |
||
| (16 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
=Overview= | =Overview= | ||
Bacteria are constantly subjected to bacteriophages and other selfish genetic elements. Bacteriophages are viruses that specifically infect bacteria and the relationship can be described as a parasitic. This is because bacteria are harmed throughout the phage replication cycle and often lysed when progeny phage particles leave the | [[File:Deleteme.jpeg|300px|thumb|right|Image 1- "E.coli" under T4 phage infection. ]] | ||
Bacteria are constantly subjected to [http://en.wikipedia.org/wiki/Bacteriophage bacteriophages] and other [http://en.wikipedia.org/wiki/Selfish_DNA selfish genetic elements]. Bacteriophages are viruses that specifically infect bacteria and the relationship can be described as a parasitic. This is because bacteria are harmed throughout the phage replication cycle and often lysed when progeny phage particles leave the cell [[#References|[1]]]. Moreover, it is estimated that there are 10^30-10^32 total phage particles on earth, which outnumber bacteria by 10-fold [[#References|[2]]]. This means that phages are found in almost every environment in which bacteria exist, making virtually all bacteria susceptible to phage infection. | |||
In response to this constant exposure to phage, bacteria have evolved several diverse antiviral defense mechanisms. These mechanisms include adsorption blocking, uptake block, abortive infection, restriction modification and the CRISPR-Cas system [[#References|[1]]]. Restriction modification and the CRISPR-Cas system are elaborated on in this article as they are the best understood and important tools in molecular biology. | |||
=Key Defense Mechanisms= | =Key Defense Mechanisms= | ||
| Line 15: | Line 13: | ||
==Restriction Modification== | ==Restriction Modification== | ||
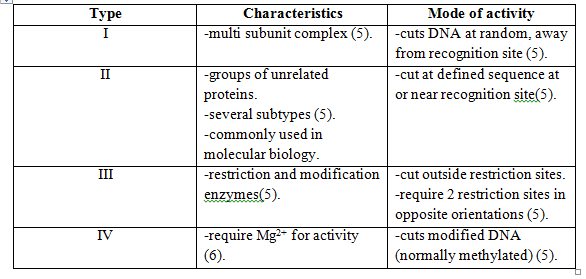
Restriction modification (R-M) is a defense mechanism which is widely spread among | Restriction modification (R-M) is a defense mechanism which is widely spread among bacteria [[#References|[3]]]. There are several types of R-M and all of these typically involve at least two enzymes, a restriction [http://en.wikipedia.org/wiki/Endonuclease endonuclease] (REase) and a [http://en.wikipedia.org/wiki/Methyltransferase methyltransferase] (MTase). The REase is responsible for the cleavage of intruding double-stranded DNA, e.g. phage genomes4, through the recognition of the specific nucleotide restriction sites. Upon genome cleavage the phage is not able to finish its life cycle. [http://en.wikipedia.org/wiki/Methylation Methylation] of restriction sites by MTase protects the host cell genome from cleavage and REases are categorized as different restriction endonuclease types depending on their specific mode of activity [[#References|[3]]]. | ||
[[File:Delete3.png| Table 1 – Comparison of types of restriction endonucleases. ]] | |||
==CRISPR== | |||
The CRISPR-Cas system is a mechanism that evolved in bacteria and archea to protect against genetic element intrusion and functions similarly to an adaptive immune system. Clustered regularly interspaced short palindromic repeats (CRISPR), are loci with several non-continuous direct repeats separated by stretches of variable sequences called spacers [[#References|[7]]] . These repeat and spacer sequences, along with one or several cas (CRISPR associated) genes, are key elements present in every CRISPR-Cas system mechanism [[#References|[7]]]. | |||
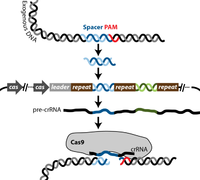
[[File:Delete2.png|200px|thumb|right| Figure 1- CRISPR-Cas type II mechanism is shown.]] | |||
There are three known types of CRISPR-Cas systems as well as several diverse subtypes. The most commonly used CRISPR-Cas system in molecular biology is type II, which is naturally found in Streptococcus thermophiles, a lactic acid bacterium that is important to the dairy industry. Cas 9 is the key enzyme required for the CRISPR system to function and has several enzymatic functions, including endonuclease and integrase activities[[#References|[7,8]]] . Cas 9 recognizes specific dsDNA sequences in the phage genome called protospacer adjacent motive (PAM), and uptakes a prospacer nucleotide sequence of about 30 bases downstream of the PAM site. This sequence is then integrated by Cas9 as a spacer into the bacterial genome flanked by two repeat sequences. The spacer sequence then gets transcribed into a precursor CRISPR RNA (pre-crRNA). The pre-crRNA gets processed by an RNase which is triggered through a transactivating crRNA (tracrRNA) that is complementary to the repeat. The process happens in the presence of Cas 9, which then associates with the processed crRNA [[#References|[8]]]. Upon re-encounter of the same phage genome, site-specific cleavage by Cas 9 occurs as the target site is determined by base complementarity between crRNA and the prospacer in the phage genome [[#References|[8]]]. | |||
=Bacteria and Phage Arms Race= | |||
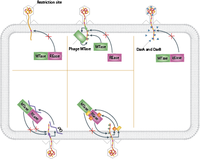
Bacteria and phage are in a constant arm race of co-evolving defense mechanisms. For example, while bacterial defense mechanisms like CRISPR and restriction modifications have evolved, phages have evolved several ways to overcome these. In terms of restriction endonucleases, there are several active and passive ways through which phage avoid cleavage. Passive mechanisms include abundance, spacing and orientation of restriction sites. Active mechanisms are more specific and in most cases include specific viral proteins have evolved to either inhibit restriction site recognition or proper REase activity [[#References|[3]]]. | |||
[[File:Phagedefense.png|200px|thumb|right| Figure 2- Graphic representing different mechanisms by which restriction enyzmes prevent phage infection. ]] | |||
To overcome the CRISPR-Cas bacterial defense, phages have evolved both simple and complex mechanisms. In certain cases, a simple point mutation in the PAM avoids acquisition of spacer sequence by Cas enzymes. Sometimes the whole prospacer and/or PAM site is deleted from the viral genome, as long as the deletion doesn’t significantly impair the phage replication cycle. Other phages harbour complex anti-CRIPSR proteins encoded in their genome. It seems like these proteins inhibit cleavage of Cas enzymes by preventing proper Cas-crRNA complex formation. Also, recent studies suggest phages have evolved a CRISPR-Cas system themselves. So far, these phage CRISPR-Cas system seem to form a Cas-crRNA in a similar fashion as the bacterial one, which can then deactivate the bacterial CRISPR defense system [[#References|[3]]]. | |||
= | =Application of Defense Systems in Biotechnology= | ||
CRISPR and restriction modification are defense mechanisms to phage infection which have vast applications in molecular biology and biotechnology. Restriction endonucleases are powerful tools in molecular biology and several specific fields, such as metabolic engineering, could have not been imagined without restriction enzymes. Restriction enzymes of the type II mechanism are the most common in laboratory applications and they effectively enable manipulation of foreign DNA through site specific cleavage [[#References|[9]]] | |||
The function of the CRISPR-Cas system is a fairly new discovery, and there are already several different applications it is used for. Most notably, the CRISPR-Cas system is an advanced and novel approach in genome engineering. The specificity of the CRISPR system allows screening for a desired mutations within genomes and occurs through crRNA:Cas directed cleavage at targeted sites [[#References|[9]]]. The CRISPR-Cas system could also be used to artificially immunize bacterial strains against specific phages. This has many potential applications in the food industry, as many processes are dependent on bacteria, such as the dairy industry. In such industries, engineered phage immunity could decrease large economic losses that are caused by phage mediated infections [[#References|[10]]]. The CRISPR-Cas has many applications, and the future of this site specific nuclease will undoubtedly provide much more biotechnological advancement in the future. | |||
=References= | =References= | ||
(1) | (1) Westra, E. R. et al. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 46, 311–39 (2012). | ||
(2) Marcó, M. B., Moineau, S. & Quiberoni, A. Bacteriophages and dairy fermentations. Bacteriophage 2, 149–158 (2012). | |||
(2) | (3) Samson, J. E., Magadán, A. H., Sabri, M. & Moineau, S. Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–87 (2013). | ||
(4) Szalay, A. A., Mackey, C. J. & Langridge, W. H. R. Restriction endonucleases and their applications JlJIIEIE. 1, 154–164 (1979). | |||
(3) | (5) Types of Restriction Endonucleases. | ||
(6) Williams, R. J. Restriction Endonucleases. 23, (2003). | |||
(4) | (7) Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–12 (2007). | ||
(8) Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–21 (2012). | |||
(5) | (9) Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L. a. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–9 (2013). | ||
(10) Brüssow, H. Phages of Dairy bacteria. 283–303 (2001). | |||
(6) | |||
(7) | |||
(8) | |||
(9) | |||
(10) | |||
Latest revision as of 02:34, 30 November 2013
Overview
Bacteria are constantly subjected to bacteriophages and other selfish genetic elements. Bacteriophages are viruses that specifically infect bacteria and the relationship can be described as a parasitic. This is because bacteria are harmed throughout the phage replication cycle and often lysed when progeny phage particles leave the cell [1]. Moreover, it is estimated that there are 10^30-10^32 total phage particles on earth, which outnumber bacteria by 10-fold [2]. This means that phages are found in almost every environment in which bacteria exist, making virtually all bacteria susceptible to phage infection.
In response to this constant exposure to phage, bacteria have evolved several diverse antiviral defense mechanisms. These mechanisms include adsorption blocking, uptake block, abortive infection, restriction modification and the CRISPR-Cas system [1]. Restriction modification and the CRISPR-Cas system are elaborated on in this article as they are the best understood and important tools in molecular biology.
Key Defense Mechanisms
Restriction Modification
Restriction modification (R-M) is a defense mechanism which is widely spread among bacteria [3]. There are several types of R-M and all of these typically involve at least two enzymes, a restriction endonuclease (REase) and a methyltransferase (MTase). The REase is responsible for the cleavage of intruding double-stranded DNA, e.g. phage genomes4, through the recognition of the specific nucleotide restriction sites. Upon genome cleavage the phage is not able to finish its life cycle. Methylation of restriction sites by MTase protects the host cell genome from cleavage and REases are categorized as different restriction endonuclease types depending on their specific mode of activity [3].
CRISPR
The CRISPR-Cas system is a mechanism that evolved in bacteria and archea to protect against genetic element intrusion and functions similarly to an adaptive immune system. Clustered regularly interspaced short palindromic repeats (CRISPR), are loci with several non-continuous direct repeats separated by stretches of variable sequences called spacers [7] . These repeat and spacer sequences, along with one or several cas (CRISPR associated) genes, are key elements present in every CRISPR-Cas system mechanism [7].
There are three known types of CRISPR-Cas systems as well as several diverse subtypes. The most commonly used CRISPR-Cas system in molecular biology is type II, which is naturally found in Streptococcus thermophiles, a lactic acid bacterium that is important to the dairy industry. Cas 9 is the key enzyme required for the CRISPR system to function and has several enzymatic functions, including endonuclease and integrase activities[7,8] . Cas 9 recognizes specific dsDNA sequences in the phage genome called protospacer adjacent motive (PAM), and uptakes a prospacer nucleotide sequence of about 30 bases downstream of the PAM site. This sequence is then integrated by Cas9 as a spacer into the bacterial genome flanked by two repeat sequences. The spacer sequence then gets transcribed into a precursor CRISPR RNA (pre-crRNA). The pre-crRNA gets processed by an RNase which is triggered through a transactivating crRNA (tracrRNA) that is complementary to the repeat. The process happens in the presence of Cas 9, which then associates with the processed crRNA [8]. Upon re-encounter of the same phage genome, site-specific cleavage by Cas 9 occurs as the target site is determined by base complementarity between crRNA and the prospacer in the phage genome [8].
Bacteria and Phage Arms Race
Bacteria and phage are in a constant arm race of co-evolving defense mechanisms. For example, while bacterial defense mechanisms like CRISPR and restriction modifications have evolved, phages have evolved several ways to overcome these. In terms of restriction endonucleases, there are several active and passive ways through which phage avoid cleavage. Passive mechanisms include abundance, spacing and orientation of restriction sites. Active mechanisms are more specific and in most cases include specific viral proteins have evolved to either inhibit restriction site recognition or proper REase activity [3].
To overcome the CRISPR-Cas bacterial defense, phages have evolved both simple and complex mechanisms. In certain cases, a simple point mutation in the PAM avoids acquisition of spacer sequence by Cas enzymes. Sometimes the whole prospacer and/or PAM site is deleted from the viral genome, as long as the deletion doesn’t significantly impair the phage replication cycle. Other phages harbour complex anti-CRIPSR proteins encoded in their genome. It seems like these proteins inhibit cleavage of Cas enzymes by preventing proper Cas-crRNA complex formation. Also, recent studies suggest phages have evolved a CRISPR-Cas system themselves. So far, these phage CRISPR-Cas system seem to form a Cas-crRNA in a similar fashion as the bacterial one, which can then deactivate the bacterial CRISPR defense system [3].
Application of Defense Systems in Biotechnology
CRISPR and restriction modification are defense mechanisms to phage infection which have vast applications in molecular biology and biotechnology. Restriction endonucleases are powerful tools in molecular biology and several specific fields, such as metabolic engineering, could have not been imagined without restriction enzymes. Restriction enzymes of the type II mechanism are the most common in laboratory applications and they effectively enable manipulation of foreign DNA through site specific cleavage [9]
The function of the CRISPR-Cas system is a fairly new discovery, and there are already several different applications it is used for. Most notably, the CRISPR-Cas system is an advanced and novel approach in genome engineering. The specificity of the CRISPR system allows screening for a desired mutations within genomes and occurs through crRNA:Cas directed cleavage at targeted sites [9]. The CRISPR-Cas system could also be used to artificially immunize bacterial strains against specific phages. This has many potential applications in the food industry, as many processes are dependent on bacteria, such as the dairy industry. In such industries, engineered phage immunity could decrease large economic losses that are caused by phage mediated infections [10]. The CRISPR-Cas has many applications, and the future of this site specific nuclease will undoubtedly provide much more biotechnological advancement in the future.
References
(1) Westra, E. R. et al. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 46, 311–39 (2012). (2) Marcó, M. B., Moineau, S. & Quiberoni, A. Bacteriophages and dairy fermentations. Bacteriophage 2, 149–158 (2012). (3) Samson, J. E., Magadán, A. H., Sabri, M. & Moineau, S. Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–87 (2013). (4) Szalay, A. A., Mackey, C. J. & Langridge, W. H. R. Restriction endonucleases and their applications JlJIIEIE. 1, 154–164 (1979). (5) Types of Restriction Endonucleases. (6) Williams, R. J. Restriction Endonucleases. 23, (2003). (7) Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–12 (2007). (8) Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–21 (2012). (9) Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L. a. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–9 (2013). (10) Brüssow, H. Phages of Dairy bacteria. 283–303 (2001).