Bdellovibrio exovorus: Difference between revisions
| Line 30: | Line 30: | ||
a. <I>Bdellovibrio exovorus</i> is a parasitic, predatory bacterium whose name means “Leech-like vibrating outside-devourer” (Gr. <I>Bdella</I>, leech, sucker; L. <I>vibrio</I>, vibrating; Gr. <I>exo-</I>, outside, L. <I>-vorus</I>, to eat.) (2). <I>B. exovorus</I> was first isolated from sewage in London, Ontario, Canada, and sequenced in 2013 by the Weizmann Institute of Science (3). | a. <I>Bdellovibrio exovorus</i> is a parasitic, predatory bacterium whose name means “Leech-like vibrating outside-devourer” (Gr. <I>Bdella</I>, leech, sucker; L. <I>vibrio</I>, vibrating; Gr. <I>exo-</I>, outside, L. <I>-vorus</I>, to eat.) (2). <I>B. exovorus</I> was first isolated from sewage in London, Ontario, Canada, and sequenced in 2013 by the Weizmann Institute of Science (3). | ||
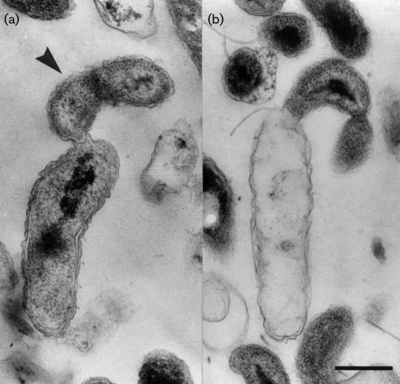
b. <I>B. exovorus</I> are gram-negative, comma-shaped rods, about 0.5 um wide and 0.5-1.4 um long. They have a single, polar, sheathed flagellum that is 29 nm wide. These | b. <I>B. exovorus</I> are gram-negative, comma-shaped rods, about 0.5 um wide and 0.5-1.4 um long. They have a single, polar, sheathed flagellum that is 29 nm wide. These obligate predators from small plaques on lawns of prey bacteria. <I>B. exovorus'</I> most common prey cell is <I>Caulobacter crescentus,</I> and actually will not prey upon <I>Escherichia coli</I> (2). | ||
Revision as of 03:06, 1 April 2017
Classification
Higher order taxa
Domain: Bacteria
Phylum: Proteobacteria
Class: Deltaproteobacteria
Order: Bdellovibrionales
Family: Bdellovibrionaceae
Species
|
NCBI: Taxonomy [1] |
Bdellovibrio exovorus
Description and significance
a. Bdellovibrio exovorus is a parasitic, predatory bacterium whose name means “Leech-like vibrating outside-devourer” (Gr. Bdella, leech, sucker; L. vibrio, vibrating; Gr. exo-, outside, L. -vorus, to eat.) (2). B. exovorus was first isolated from sewage in London, Ontario, Canada, and sequenced in 2013 by the Weizmann Institute of Science (3).
b. B. exovorus are gram-negative, comma-shaped rods, about 0.5 um wide and 0.5-1.4 um long. They have a single, polar, sheathed flagellum that is 29 nm wide. These obligate predators from small plaques on lawns of prey bacteria. B. exovorus' most common prey cell is Caulobacter crescentus, and actually will not prey upon Escherichia coli (2).
Existing genus MicrobeWiki Link: https://microbewiki.kenyon.edu/index.php/Bdellovibrio
Genome and genetics
a. Bdellovibrio exovorus belongs to the major branch of prokaryotes of deltaproteobacteria . The most closely related species is Bdellovibrio bacteriovorus . Based on 16S rRNA sequences, the most closely related genera include Desulfomonile, Desulfuromonas, and more distantly, Bacteriovorax.
b. Briefly describe any extra-chromosomal elements or genetic tools that are used to study the bacterium: viruses, plasmids, transposons that allow genetic manipulation and analysis.
Both PCR and Southern blotting with a DIG-labelled probe (DIG is Digoxigenin, a steroid only found in plants) on the hit locus (short for "host interaction"). The CRISPR system has been used to count DNA scaffolds in the genome.
c. The sequence of Bdellovibrio exovorus was determined using whole genome sequencing (Weizmann Institute of Science, 2013), and can be found in its entirety on the Joint Genome Institute database. It has a total genome size of 2,657,893 bases, a %G+C content of 41.92%, and 2,656 genes. Of those genes, 2,619 (98.61%) are protein coding genes, 37 (1.39%) are RNA genes (of which 34 are for tRNA and 3 are for rRNA), and 1 (0.04%) is a pseudogene. Its 16S rRNA gene is 1502 bp long, and is only 90-93% similar to other strains of Bdellovibrio bacteriovorus.
Nutrition and metabolism
a.Describe the growth characteristics of your bacterial species; sources of C, N, electrons; respires/ferments, uses O2, etc.
Bdellovibrio exovorus is an obligate predator, preferring to feed on bacterial species Caulobacter crescentus. From its genome, it is predicted to be auxotrophic for the metabolism of almost every amino acid, except for glutamate, glycine, and asparagine, meaning the rest must be broken down from their prey bacteria.
Bdellovibrio exovorus has a biphasic life cycle. First, a free-swimming phase where the cell attaches to the outside of a prey cell. All other known plaque-forming species within the family Bdellovibrionaceae are identified by their growth within the periplasmic space of their prey bacteria. However, Bdellovibrio exovorus has no periplasmic phase in its life cycle. Because Bdellovibrio exovorus does not inhabit the periplasmic space, it does not form the rounded bdelloplast out of the host cell that is common among members of the genus Bdellovibrio, or even related genera like Bacteriovorax, Bacteriolyticum, and Predibacter.
Replication in Bdellovibrio exovorus occurs via binary fission, with no sign of the long filamentous extracellular structures common with related species.
b.What kinds of culture conditions (temp, pH, media) are needed for laboratory study?
For laboratory study, cultures of Bdellovibrio exovorus are grown on minimal medium with the appropriate prey bacterium grown as a lawn. Bdellovibrio exovorus will grow plaques on that bacterial lawn, leaving the empty husks of the prey cells behind.
One culturing method used by researchers involved growing Bdellovibrio exovorus on 1/10-strength yest extract-peptone medium supplemented with calcium, although dilute nutrient broth medium has also been used. HM buffer is often added to media, made of HEPES, CaCl2, and MgSO4. While optimal growth occurs around 30 °C, Bdellovibrio exovorus can be kept in a sterile tube at 4 °C for 1 month or stored long-term with 25% (w/v) glycerol at -80 °C. One research team calculated a 46.1 mol% G+C content in 1x SSC (Saline-Sodium Citrate buffer) using the equation [G+C=1.99(Tm-66.0)], so its genomic DNA must have undergone thermal denaturation at 89.2 °C.
Unlike some halophilic strains of Bdellovibrio, this species prefers a less saline environment.
c.What kinds of waste, by-products, volatile compounds are generated?
Ecology / Pathology
Ecology: How is your microorganism important in the ecosystem where it is found? How does it impact other organisms in the environment (could be positive or negative impact)?
Pathology: How does the microbe cause disease as it interacts with the host? Describe any specific toxins or pathways that are used for invading and causing disease in the host. What treatment is used to inhibit or kill the microbe?
Current Research
Describe recent research and findings that have been done with this organism. The research can be clinical, applied or basic research. This section should be based on 2 recent papers (10 years or less) and summarized in your own words.
References
[2] Koval SF, Hynes SH, Flannagan RS, Pasternak Z, Davidov Y, Jurkevitch E. Bdellovibrio exovorus sp. nov., a novel predator of Caulobacter crescentus. Int. J. Syst. Evol. Microbiol [Internet]. 2013 [cited 13 Feb 2017];(63):146–151.
[3] Bdellovibrio exovorus JSS [internet]. University of California- Integrated Microbial Genomes & Microbial Samples. [cited 2017 Feb 13] Availible from https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=2541047022
[4] Brenner DJ. 2005. Bergey's manual of systematic bacteriology. Vol. 2. The proteobacteria. Part C. The alpha-, beta-, delta-, and epsilonproteobacteria. Staley JT, editor. New York (NY): Springer. p.1040-1053.
Authored by Kip Pierce, a student of CJ Funk at John Brown University