Blautia Hydrogenotrophica: Difference between revisions
| Line 15: | Line 15: | ||
==Description and Significance== | ==Description and Significance== | ||
''Blautia hydrogenotrophica'', formerly known as ''Ruminococcus hydrogenotrophicus'', is a species which inhabits the guts of mammals (Bernalier et al.). Microbial gut communities play an important role in breaking down components of the host diet which would otherwise be indigestible, mainly plant matter, thus having an impact on the host's metabolic efficiency (Bain et al.) | ''Blautia hydrogenotrophica'', formerly known as ''Ruminococcus hydrogenotrophicus'', is a species which inhabits the guts of mammals (Bernalier et al.). Microbial gut communities play an important role in breaking down components of the host diet which would otherwise be indigestible, mainly plant matter, thus having an impact on the host's metabolic efficiency (Bain et al.). | ||
==Genome Structure== | ==Genome Structure== | ||
Revision as of 13:20, 24 April 2011
Classification
Bacteria; Firmicutes; Clostridia; Clostridiales; Blautia
Species
Blautia hydrogenotrophica
|
NCBI: Taxonomy |
Description and Significance
Blautia hydrogenotrophica, formerly known as Ruminococcus hydrogenotrophicus, is a species which inhabits the guts of mammals (Bernalier et al.). Microbial gut communities play an important role in breaking down components of the host diet which would otherwise be indigestible, mainly plant matter, thus having an impact on the host's metabolic efficiency (Bain et al.).
Genome Structure
Blautia Hydrogenotrophica strain DSM 10507 contains 3,565,428 base pairs (NCBI, 2011). The G+C content of the DNA is 45.2 mol% (Bernalier et al. 1996). Through in silico analysis of the Blautia hydrogenotrophica genome, it was found that all of the genes involved in the Wood-Ljungdahl pathway were present. This includes eight genes that are thought to encode subunits of iron-only hydrogenases. (Bain, 2010)
Cell Structure, Metabolism and Life Cycle
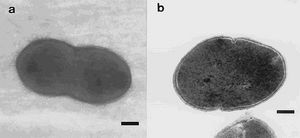
The Blautia hydrogenotrophica is gram-positive, non-sporulating, coccobacillus shaped bacterium with an average size of 0.7-0.6 mm. Through the use of negatively stained cells, it can be observed that the organism lacks flagella (Bernalier et al. 1996). Hydrogenotrophica, meaning feeds on hydrogen, refers to the organism’s ability to use H2 and CO2 as energy source for growth” (Liu et al., 2008). ‘’Blautia hydrogenotrophica’’ grows autotrophically by metabolizing H2 and CO2 to form acetate as sole metabolite. The organism is also able to grow heterotrophically using several different organic compounds as a substrate. Through the fermentation of glucose and fructose, acetate primarily produced, however ethanol, lactate and, to a lesser extent, isobutyrate and isovalerate can also be formed. (Bernalier et al. 1996)
The production of acetate from H2 and CO2 occurs through the acetyl-CoA pathway. Acetogens, such as Blautia hydrogenotrophica, have the ability to use a broad range of substrates, including mono-, di-, tri-, and tetrasaccharides, in addition to aromatic and aliphatic amino acids. By targeting aliphatic and aromatic amino acids, the efficiency of fermentation is increased. This is accomplished through the consumption of reducing equivalents, thus a high NAD+/NADH ratio is maintained and acetate production increases. Due to the variety of substrates that can be used, it is hypothesized that acetogens possess an advantage in fitness when competing for H2 and formate over methanogens and sulfate reducing bacteria, other groups of microorganisms found in the gut. Further support of this hypothesis comes from studies indicating acetogens’ ability to grow mixotrophically, meaning that organic substrates and H2 are used as an energy source simultaneously. This indicates that Blautia hydrogenotrophica would have a greater advantage over H2-consuming methanogens. (Bain et al., 2010)
Ecology
References
Bain, J., Faith, J., Gordon, J., Muehlbauer, M., Newgard, C., Rey, F., and Stevens, R. “Dissecting the in Vivo Metabolic Potential of Two Human Gut Acetogens.” Journal of Biological Chemistry, 2010. Volume 285. P. 22082-22090. Accessed April 18, 2011. <http://www.jbc.org/content/285/29/22082.full>
Bernalier, A., Collins, MD., Leclerc, M., Rochet, V., Willems, A. “Ruminococcus hydrogenotrophicus sp. nov., a new H2/CO2-utilizing acetogenic bacterium isolated from human feces.” Archives of Microbiology, 1996. Volume 166(3). p. 176-83. Accessed April 16, 2011. < http://www.springerlink.com/content/tt7hew38ngt4j9d7/>
Liu, C., Finegold, S., Song, Y., and Lawson, P. “Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces.” International Journal of Systematic and Evollutionary Microbiology, 2008. Volume 58. p. 1896-1902. Accessed April 21, 2011. <http://ijs.sgmjournals.org/cgi/content/full/58/8/1896>
NCBI (National Center for Biotechnology Information). “Blautia hydrogenotrophica DSM 10507, whole genome shotgun sequence.” Accessed April 21, 2011.
<http://www.ncbi.nlm.nih.gov/nuccore/NZ_ACBZ00000000>
Author
Page authored by Megan Buhl and Blaze Budd, students of Prof. Jay Lennon at Michigan State University.
