Bluetongue Virus (BTV)
Bluetongue Virus (BTV)
A Viral Biorealm page on the family Bluetongue Virus (BTV)
Baltimore Classification
Group III: Double-Stranded RNA Viruses
Higher order categories
Order: Unassigned
Family: Reoviridae
Subfamily: Sedoreovirinae
Genus: Orbivirus
Description and Significance
Bluetongue Virus (BTV) is a pathogenic virus that causes serious disease in livestock. BTV was first observed in Africa in the late 18th century, but has since been observed in Australia, the United States, Africa, the Middle East, Asia, and Europe. There are 24 distinct serotypes of BTV. BTV causes Bluetongue disease in ruminant species, such as sheep, goats, deer, and cattle, although clinical signs of infection are almost solely restricted to sheep [5].
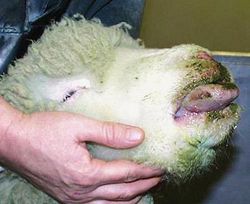
The incubation period for BTV is 5-20 days. BTV infection first manifests itself with the onset of a high fever that may last several days. The virus mainly affects small blood vessels, causing hemorrhaging, hyperemia, and edema in the tissue of the lips, mouth, nasal linings, and eyelids. Swelling of the lips and tongue, accompanied by viral destruction of host blood vessels decreases the amount of oxygen that reaches the tongue tissue. Reduced oxygen delivery causes cyanosis, or the blue appearance of the tongue. However, cyanosis is usually confined to the minority of Bluetongue cases [6].

The mortality rate of especially susceptible breeds of sheep can be as high as 70%, as a result of secondary bacterial infections. Furthermore, the high fever experienced by infected sheep can lead to wool breaks, decreasing productivity [2]. Some species, primarily cattle and goats, are asymptomatic upon BTV infection, but their productivity as livestock can be significantly reduced, with milk yields dropping nearly 40% [7]. BTV has been considered a possible bioweapon because of the difficulty associated with controlling and eliminating the disease, as well as the significant economic loss that would result from widespread infection [8].
There are no treatment options for BTV. Immunization has proved the most effective way to prevent bluetongue outbreaks in endemic regions. In southern Africa and other affected regions, a vaccine comprised of 5 attenuated BTV serotypes is widely used. In the United States, a vaccine containing a modified live version of a single serotype is utilized [9].
Genome Structure
The genome of BTV consists of a strand of linear dsRNA, 19,200 base pairs in length. The dsRNA genome can be split into 10 segments, each fragment encoding a single viral protein. The size of each segment ranges from 822 to 3954 base pairs in length. Fragment 10 is the only fragment which encodes two proteins, NS3 and NS3A, although both proteins are highly related and are the result of alternative initiation [10].
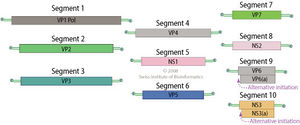
Protein-coding dsRNA fragments are translated by RNA-dependent RNA polymerase, which is encoded by VP1, segment 1 of the dsRNA genome [11]. Segments 2 and 6 produce VP2 and VP5, respectively, which form the outer layer of the capsid. Segment 7 encodes VP7, which forms the core-surface layer. Segment 3 produces VP3, the protein that forms the subcore shell. Segments 4 and 9 produce VP4 and VP6, respectively. VP4 forms an mRNA capping enzyme, while VP6 yields viral helicase. In conjunction with the polymerase produced by VP1, VP4 and VP6 form transcriptase complexes [12]. Segment 8 encodes NS2, which forms important viral inclusion body matrix proteins [13]. Viral proteins NS3 and NS3A form viral membrane proteins[14] and glycoproteins[15].
Virion Structure of BTV
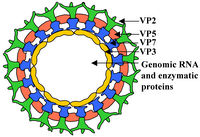
BTV virions display an icosahedral capsid enclosing a dsRNA genome. It consists of a two layer, 470S core particle, containing the dsRNA genome and integral proteins, and an outer capsid protein layer. The core particle surrounding the genome is 700 Å in length, and is the first highly complex Virion structure ever solved [16]. The outermost core layer is composed of 260 VP7 trimers. The core particle’s innermost layer consists of 60 VP3 dimers. Within the VP3 layer, transcriptional complexes of VP1 (RNA dependent RNA-polymerase), VP4 (capping enzyme) and VP6 (helicase) are found [17]. The outermost layer of the viral capsid is composed of 60 VP2 trimers and 120 VP5 trimers . These outer capsid proteins are involved in virion attachment to host cell membrane and host cell entry [18].
Reproductive Cycle of BTV in a Host Cell
The reproductive cycle of BTV is very poorly understood. The Reoviridae family of viruses lack an effective model for experimental manipulation, and therefore, progress in understanding the mechanisms of host cell invasion, viral replication and progeny exit in viruses such as BTV has been enormously challenging.
However, it is understood that outer capsid proteins VP2 and VP5 are involved in cell attachment and penetration. After the host cell is infected, BTV is uncoated, and the 470S core particle begins the viral replication cycle via the VP1-VP4-VP6 transcriptional complex. Nonstructural viral proteins NS1 and NS2 have been found in high concentrations in host cytoplasm, and therefore it is believed that they may play a role in virus replication and progeny assembly. The mechanism in which progeny exit the host cell is also poorly understood, although some researchers believe nonstructural viral protein NS3A may be involved in progeny exit [19].
Viral Ecology & Pathology
References
Example:
Weir, Jerry P. " Genomic Organization and Evolution of the Human Herpesviruses." Virus Genes 16.1 (1998): 85-93.
Page authored for BIOL 375 Virology, September 2010
