Bovine Leukemia Virus
{Uncurated}}
Classification
Viruses; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae
Species
Deltaretrovirus bovine leukemia virus (BLV)
|
NCBI:11901 [1] |
Description and Significance
D. bovine leukemia virus was originally HTLV-BLV and grouped with T-lymphotropic virus group HTLV until 1999 when it was given its own species designation [14].
- bovine leukemia virus is an oncogenic, B-lymphotropic virus that can cause Bovine Leukemia in cattle and found in a high percentage of dairy and beef cattle world-wide, with exception to some Western European countries [17].
- In the U.S. it is estimated to cause annual financial losses of ~$525 million from decreased milk production even in asymptomatic dairy cattle [16]
- Antibodies against BLV provirus, Viral DNA in healthy and non-healthy breast tissue and a correlation with cancerous breast tumors have all been found in humans [16]
- Mechanisms behind BLV leading to Bovine Leukemia can be studied to better understand pathogenesis of human T-cell leukemia virus
Genome Structure
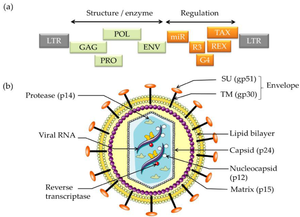
The BLV retrovirus is a diploid, single-stranded RNA virus that through reverse transcription becomes a double-stranded DNA provirus. Different isolated strains of BLV have been sequenced 154 times since 1997, with varying number of nucleotides from 7,933 to 8,714. In 2019 a complete genome with 8,419 nucleotides and GC content of 54.16% was accepted as a representative Deltaretrovirus bovine leukemia virus genome [14]. The viral genome contains the 6 protein coding genes and accessory genes shown below [17].
- Genome Structure
- gag - codes for matrix protein, capsid protein and nucleocapsid protein (p12, p24 & p15)
- pro - codes protease (Pro)
- pol - codes reverse transcriptase & integrase (RT & IN)
- env - mature extracellular protein and transmembrane protein (gp51 & gp30)
- R3 & G4 - accessory genes
- MicroRNAs - between 'env' and pX region are the RNA polymerase-III-encoded MicroRNAs
- tax & rex - regulatory proteins (pX region)
Through whole genome and env gene sequencing, 10 - 11 genotypes have been identified. Many are named according to the geographic region they are most prevalent or originated it. However genotypes 1-3 are found throughout all regions where the virus is found [17].
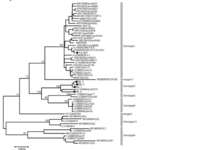
- Researchers sequencing the provirus env gene constructed the phylogenic tree found in fig.1 [15]
- Stable provirus genome with sequenced strains sharing 94.8-99.5% similarity [15]
- Genomic sequences available include the isolate organisms named according to geographic region listed below [15].
- -Isolate organisms::American Isolate FLK; American Isolate VDM; Australian Isolate; Belgium Isolate LB285; Belgium Isolate LB59; Japanese Isolate BLV-1.
Cell Structure, Metabolism and Life Cycle
Cell Structure The mature BLV virion structure encodes structural enzymes GAG, POL, PRO, and ENV. Regulation is encoded by miR, R3, G4, REX, and TAX. The external section is an envelope containing glycoproteins SU (gp51 and TM). Moving into the cell is the lipid bilayer and matrix. Inside the virus, the reverse transcriptase, viral RNA, and nucleocapsid are protected by the capsid (p24)[1].
Metabolism Viruses are non-living entities and do not have their own metabolism. However, viruses can modify the cellular metabolism of the host cells that they enter [9]. Researchers studying BLV are interested in the TAX gene's role in viral metabolism. The TAX transactivator is a transcriptional activator of viral expression and contributes to viral expression. In rat studies, TAX causes immortalization of rat fibroblasts, cells found in the connective tissue of rats, and is vital for healing wounds [5]. Ha-ras is an oncogene that works with TAX to induce full transformation of cells that form tumors. TAX is a promising target because it has a very narrow range of variations and is compatible with full transactivation activity, suggesting that the present molecular structure of TAX results from heavy evolutionary constraints [2].
Life Cycle Viruses must use host cell machinery to undergo viral replication [8]. The general viral life cycle includes entering the host cell, adapting to the host’s intracellular environment, shedding its capsid, transcribing its RNA, translating its RNA to the viral proteins, replicating its genome, assembling the viral components, and exiting from the host cell. The env gene encodes gpr72, a polypeptide precursor in the BLV virus. It is cleaved into two glycoproteins: a surface protein (gp51) and a transmembrane protein (gp30)[6]. This area contains the recognition site required for viral entry and mediates cell fusion, which is necessary for viral entry into the cell. Further, the BLV provirus produces many viral microRNAs that express antisense transcripts, suggesting that they play an important role in the viral life cycle and tumorigenic potential. This is not a comprehensive overview of the BLV life cycle but outlines parts of the viral life cycle that are known to be important for viral propagation.
Ecology and Pathogenesis
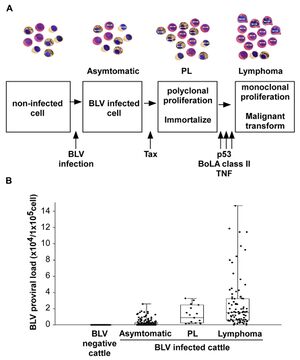
Pathogenesis The infection route of BLV is both through horizontal and vertical transmission. BLV is transmitted via direct contact (Kono et al., 1983), milk, and insect bites. One of the most common manners of infection is through blood in milk and flies. There are three main stages of BVL. The first begins with an infection, and a cell contains the virus but is asymptomatic. The steps then begin with the Tax protein, a transcriptional activator of viral gene expression. It may inhibit DNA repair mechanisms, therefore increasing mutations within the DNA,. The next stage is known as PL, polyclonal proliferation of the virus in cells (called immortalization). Although most BLV-infected cows present no symptoms, PL is a benign form of the disease that describes an increase of untransformed B-lymphocytes in the animal's blood. However, even after proliferation, it cannot transform cells. For lymphoma to develop, it needs a malignant transformation through a P53 mutant, TNF, or BoLa [1].
- P53 is a common tumor suppressor gene that works to control cell proliferation and apoptosis. Studies show that about half of tumors created by the BLV in cattle also contained missense mutations in p53. This suggests that p53 is one of many genetic changes that play a role in the further development of the disease.
- TNF, known as a tumor necrosis factor, TNF is a protein made by white blood cells in response to an antigen. It was found to be much higher in animals with lymphoma.
- BoLa is a bovine leukocyte antigen that determines the immune responsiveness of the animal. The genetic variations of amino acids influence susceptibility. An example of this is seen in the presence of the amino acids Glu–Arg that confer resistance to PL in BLV-infected cattle.
Symptoms Only 29% of BLV-infected cattle develop persistent lymphocytosis (which are benign tumors but full of infectious potential), while <5% of BLV-infected cattle develop lymphosarcoma. Symptoms include extreme weight loss, labored breathing, lesions, tumors, bloat, fever, and loss of muscle. Lymphocytosis occurs as seen in PL, which increases B-lymphocytes past the normal range.
Significance to the Microbiome Studies show that the fecal microbial diversity differed slightly between affected and unaffected cows, and bacterial taxa associated with rumen fermentation were more common in unaffected cows. The diversity of the microbiota was also affected, and more proliferation of BLV led to less diversity [11]. The rumen of the cow is responsible for breaking down food into useable energy and protein, so when BLV modifies the rumen's energy efficiency of this system, it causes negative effects. This may explain lessened milk production and susceptibility to other illnesses when infected with BLV.
References
[1]Aida, Yoko, et al. “Mechanisms of Pathogenesis Induced by Bovine Leukemia Virus as a Model for Human T-Cell Leukemia Virus.” Frontiers in Microbiology, vol. 4, 2013, https://doi.org/10.3389/fmicb.2013.00328.
[2] Barez, P. Y., de Brogniez, A., Carpentier, A., Gazon, H., Gillet, N., Gutiérrez, G., Hamaidia, M., Jacques, J. R., Perike, S., Neelature Sriramareddy, S., Renotte, N., Staumont, B., Reichert, M., Trono, K., & Willems, L. (2015). Recent Advances in BLV Research. Viruses, 7(11), 6080–6088. https://doi.org/10.3390/v7112929
[3]Bovine Leukemia Virus Vaccine. Creative Biolabs Vaccine. https://www.creative-biolabs.com/vaccine/bovine-leukemia-virus-vaccine.htm
[4]Deltaretrovirus. ViralZone (SIB Swiss Institue of Bioinformatics). https://viralzone.expasy.org/91
[5]Gillet, N., Florins, A., Boxus, M., Burteau, C., Nigro, A., Vandermeers, F., Balon, H., Bouzar, A. B., Defoiche, J., Burny, A., Reichert,
[6]M., Kettmann, R., & Willems, L. (2007). Mechanisms of leukemogenesis induced by bovine leukemia virus: prospects for novel anti-retroviral therapies in human. Retrovirology, 4, 18. https://doi.org/10.1186/1742-4690-4-18
[7]Marawan, M. A., Alouffi, A., El Tokhy, S., Badawy, S., Shirani, I., Dawood, A., Guo, A., Almutairi, M. M., Alshammari, F. A., & Selim, A. (2021). Bovine Leukaemia Virus: Current Epidemiological Circumstance and Future Prospective. Viruses, 13(11), 2167. https://doi.org/10.3390/v13112167
[8]Moratorio, G., Fischer, S., Bianchi, S. et al. (2013). A detailed molecular analysis of complete Bovine Leukemia Virus genomes isolated from B-cell lymphosarcomas. Vet Res 44, 19 . https://doi.org/10.1186/1297-9716-44-19
[9]Ryu W. S. (2017). Virus Life Cycle. Molecular Virology of Human Pathogenic Viruses, 31–45. https://doi.org/10.1016/B978-0-12-800838-6.00003-5
[10]Sanchez, E., Lagunoff, M. (2015). Viral activation of cellular metabolism. Virology 470-408, 609-618. https://doi.org/10.1016/j.virol.2015.02.038.
[11]Uchiyama, Jumpei et al. “Examination of the fecal microbiota in dairy cows infected with bovine leukemia virus.” Veterinary microbiology vol. 240 (2020): 108547. doi:10.1016/j.vetmic.2019.108547
[12]Yoko, A., Murakami, H., Takahashi, M., Takeshima, S. (2013). Mechanisms of pathogenesis induced by bovine leukemia virus as a model for human T-cell leukemia virus. Frontiers in Microbiology 4,https://doi.org/10.3389/fmicb.2013.00328
[13] Schoch CL, et al. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020: baaa062. PubMed: 32761142 PMC: PMC7408187.
[14] Lefkowitz, E. J., Dempsey, D. M., Hendrickson, R. C., Orton, R. J., Siddell, S. G., & Smith, D. B. (2018). Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Research, 46(D1), D708–D717. https://doi.org/10.1093/nar/gkx932
[15] Vafin, R. R., Khazipov, N. Z., Shaeva, A. Y., Zakirova, Z. R., Zaynullin, L. I., Tyulkin, S. V., Abdulina, I. R., & Alimov, A. M. (2014). Genotypic identification of the bovine leukemia virus. Molecular Genetics, Microbiology and Virology, 29(4), 195–203. https://doi.org/10.3103/S0891416814040120
[16] Buehring, G. C., DeLaney, A., Shen, H., Chu, D. L., Razavian, N., Schwartz, D. A., Demkovich, Z. R., & Bates, M. N. (2019). Bovine leukemia virus discovered in human blood. BMC Infectious Diseases, 19(1), 297. https://doi.org/10.1186/s12879-019-3891-9
[17] Polat, M., Takeshima, S., & Aida, Y. (2017). Epidemiology and genetic diversity of bovine leukemia virus. Virology Journal, 14(1), 209. https://doi.org/10.1186/s12985-017-0876-4
Author
Page authored by Grace Williams, Ashlee Webb, Laura Gerber, students of Prof. Jay Lennon at Indiana University.