Candidatus Tremblaya princeps
Classification
Domain: Bacteria
Phylum: Pseudomonadota
Class: Betaproteobacteria
Genus: Candidatus Tremblaya
This bacterium belongs to the Betaproteobacteria class, a diverse group known for both their ecological versatility and economic significance. Within this class, Ca. T. princeps stands out due to its specialized symbiotic lifestyle and extreme genomic reduction. It is classified as a 'Candidatus' genus, indicating it has not yet been cultured in a lab setting [2].
Species
|
NCBI: [1] |
Candidatus Tremblaya princeps
Description and Significance
Background
The study of symbiotic relationships between insects and microorganisms has a history dating back to the mid-20th century, with key milestones that have significantly shaped our understanding of these intricate and fascinating associations. One of the foundational studies in this field was conducted by Paul Buchner in the 1940s, where he focused on the exploration of bacteriocytes in insects, particularly mealybugs. Buchner's work, documented in publications such as “Endosymbiosis of Animals with Plant Microorganisms” (1965), laid the groundwork for recognizing the presence of symbiotic microorganisms within specialized cells of insects, including Candidatus Tremblaya princeps.
In the 1950s, Vincent Wigglesworth and Paul Buchner made significant contributions to our understanding of insect physiology and cellular biology. Wigglesworth, known for his comprehensive work in insect physiology, including his seminal work “The Principles of Insect Physiology” (1972) focused on studying various physiological processes within insects, including nutrition, respiration, circulation, and reproduction. While his research did not directly investigate symbiotic relationships or symbiotic bacteria, his detailed observations of insect cellular structures, such as bacteriocytes, provided foundational knowledge about the cellular environments in which symbiotic relationships occur.
A landmark study titled “An interdependent metabolic patchwork in the nested symbiosis of mealybugs” by McCutcheon and von Dohlen in 2011, provided a deeper insight into the genomic structure of Candidatus Tremblaya princeps. In this study, McCutcheon and von Dohlen described the highly reduced genome of Ca. T. princeps and detailed how it relies on both the mealybug host and another bacterium, Candidatus Moranella endobia, for essential nutrients and metabolic functions. The research highlighted this extreme example of codependence in nested symbiotic relationships, showing how Candidatus Tremblaya princeps has lost many genes for vital metabolic pathways, which are instead provided by its co-symbiont, Candidatus Moranella endobia. They discovered that this bacterium has one of the smallest genomes of any known cellular organism, containing only essential genes for its symbiotic existence and relying heavily on its host for survival.
Significance
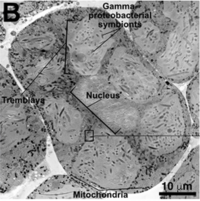
Candidatus Tremblaya princeps is primarily recognized for its nested symbiotic relationship within mealybugs, particularly the citrus mealybug Planococcus citri, where it is encapsulated in specialized bacteriocytes. It is unique for housing another endosymbiont, Moranella endobia, within its own cellular structure. This multilayered endosymbiosis is a remarkable example of evolutionary adaptation, where Ca. T. princeps, despite its extremely reduced genome, plays a critical role in the nutritional pathways of its host. Its genome is one of the smallest known among prokaryotes, reflecting significant genomic streamlining possibly due to its symbiotic lifestyle [3,4].
The small size of the genome brings about significant dependencies, where essential metabolic functions such as synthesis of amino acids and nucleotides are outsourced to its nested symbiont or the mealybug host. This makes Ca. T. princeps an interesting subject for studies on cellular minimalism and symbiotic interdependence [3,5].
Genome Structure
With a circular genome of only 138,931 base pairs, Ca. Tremblaya princeps has the second smallest genome and the smallest number of functional genes present within a prokaryote. Its genome only encodes 116 protein-coding genes and 20 RNA-coding genes and is so reduced that it no longer has a complete set of enzymes for any one pathway [2,6,8]. This reduction can be attributed to genome streamlining, which is a common occurrence with endosymbionts. T. princeps is an extreme example, however, due to its multi-tiered symbiosis. The incomplete pathways found within the T. princeps are complemented by the genomes of both the secondary endosymbiont and the mealybug, creating a three-way dependency. As a reference, Mycoplasma genitalium, which is often used for minimal cell studies and widely considered the smallest free-living prokaryote. It has a genome size of 580,076 bp and about 480 proteins, remarkably making it considerably larger than T. princeps [6,7].
Cell Structure, Metabolism and Life Cycle
Candidatus Tremblaya princeps exhibits a highly specialized cellular structure adapted to its endosymbiotic lifestyle. Lacking many basic metabolic pathways, it relies heavily on both its mealybug host and the Moranella endobia for survival and replication. The cell structure is characterized by a compact design with essential genes mainly focused on the production of amino acids and ribosomal components necessary for protein synthesis. It has retained the ability to synthesize key amino acids such as valine, leucine, and isoleucine, which are critical for the host's nutrition, particularly given the nutrient-poor sap diet of mealybugs. The bacterium is an obligate endosymbiont, meaning it cannot survive outside its host's body. Its life cycle is closely tied to that of its host, with strict vertical transmission ensuring that each generation of mealybugs carries the endosymbiont. The interdependence is so critical that the bacterium and its host have co-evolved mechanisms to ensure mutual survival and replication [2,3,5,8].

Ecology and Pathogenesis
The ecological role of Candidatus Tremblaya princeps is defined through its symbiotic relationships. Its presence within the mealybug Planococcus citri is essential for the insect's survival, particularly in nutrient-poor environments where the synthesis of essential amino acids by Ca. T. princeps and Moranella endobia is vital [8, 3]. Additionally, some strains of Tremblaya strains in a different species of mealybugs, Pseudococcus longispinus have been shown to house two distinct endosymbionts within each cell. These two secondary endosymbionts have not been identified previously, but based off of their respective closest potential relatives, they have been tentatively named Candidatus Sodalis endolongispinus and Candidatus Symbiopectobacterium endolongispinus [1].
The nested symbiotic relationships that T. princeps participates in allow for a remarkable efficiency in nutrient production and conservation, enabling mealybugs to thrive on sap diets that lack many essential nutrients. The study of Ca. T. princeps offers insights into the evolutionary dynamics of genome reduction and the ecological and evolutionary implications of such extreme symbiotic relationships. Understanding this symbiosis can also contribute to more effective biological control strategies for managing mealybug populations, which are significant pests in agriculture.
References
[1] Arkadiy I Garber, Maria Kupper, Dominik R Laetsch, Stephanie R Weldon, Mark S Ladinsky, Pamela J Bjorkman, John P McCutcheon. 2021. The Evolution of Interdependence in a Four-Way Mealybug Symbiosis. Genome Biology and Evolution 13(8): evab123. https://doi.org/10.1093/gbe/evab123
[2] López-Madrigal S, Latorre A, Porcar M, Moya A, Gil R. 2011. Complete genome sequence of "Candidatus Tremblaya princeps" strain PCVAL, an intriguing translational machine below the living-cell status. J Bacteriol 193(19):5587-8. doi:10.1128/JB.05749-11
[3] López-Madrigal S, Balmand S, Latorre A, Heddi A, Moya A, Gil R. 2013. How does Tremblaya princeps get essential proteins from its nested partner Moranella endobia in the Mealybug Planoccocus citri? ‘’PLoS One’’ 8(10):e77307. doi:10.1371/journal.pone.0077307
[4] López-Madrigal S, Latorre A, Moya A, Gil R. 2015. The link between independent acquisition of intracellular gamma-endosymbionts and concerted evolution in Tremblaya princeps. ’’Front Microbiol’’ 6:642. doi:10.3389/fmicb.2015.00642
[5] López-Madrigal S, Beltrà A, Resurrección S, Soto A, Latorre A, Moya A, Gil R. 2014. Molecular evidence for ongoing complementarity and horizontal gene transfer in endosymbiotic systems of mealybugs. ‘’Front Microbiol’’ 5:449. doi:10.3389/fmicb.2014.00449
[6] Martínez-Cano DJ, Reyes-Prieto M, Martínez-Romero E, Partida-Martínez LP, Latorre A, Moya A, Delaye L. 2015. Evolution of small prokaryotic genomes. ‘’Front Microbiol’’ 5:742. doi: [10.3389/fmicb.2014.00742](https://doi.org/10.3389/fmicb.
[7] McCutcheon J, Moran N. 2012. Extreme genome reduction in symbiotic bacteria. ‘’Nat Rev Microbiol’’ 10:13–26. https://doi.org/10.1038/nrmicro2670
[8] McCutcheon JP, von Dohlen CD. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. ‘’Curr Biol’’ 21(16):1366-72. doi:10.1016/j.cub.2011.06.051
Author
Page authored by Alex Ogden and El Park, students of Prof. Jay Lennon at Indiana University.