Chlorobium FMO antenna complex characterisation: Difference between revisions
| Line 23: | Line 23: | ||
In ''C. tepidum'', the Fenna-Matthews-Olson protein is part of a light-harvesting complex that consists of three major components: 1. Chlorosome 2. Aforementioned FMO protein, and 3. Reaction center. This system captures electromagnetic radiation from the sun and forms a funnel-like system that directs energy transfer (3). The role of the FMO protein is analogous to that of a wire connecting two electrodes. That is to say that the FMO protein provides the transfer of energy from the chlorosome to the reaction center. | In ''C. tepidum'', the Fenna-Matthews-Olson protein is part of a light-harvesting complex that consists of three major components: 1. Chlorosome 2. Aforementioned FMO protein, and 3. Reaction center. This system captures electromagnetic radiation from the sun and forms a funnel-like system that directs energy transfer (3). The role of the FMO protein is analogous to that of a wire connecting two electrodes. That is to say that the FMO protein provides the transfer of energy from the chlorosome to the reaction center. | ||
[[Image:Light Harvesting Model of Green Sulfur Bacteria 1.png|frame|left|Model depicting the process of light harvesting in C. tepidum. The model includes the chlorosome, FMO protein, and reaction center. Photo courtesy of the University of Copenhagen Biology Department (http://www.bio.ku.dk/nuf/images/Ctepmodel040107_mod.jpg)]] | |||
Include some current research in each topic, with at least one figure showing data. | Include some current research in each topic, with at least one figure showing data. | ||
Revision as of 06:02, 15 April 2009
Discovery of the Fenna-Matthews-Olson Protein Membrane Orientation in Chlorobaculum tepidum
A team of researchers at Washington University in St. Louis, lead by Robert Blankenship, has pioneered a new method of discovering protein orientation in living systems (1). By combining chemical labeling with mass spectroscopy, these scientists brought forth knowledge of the structure/function relationship of the Fenna-Matthews-Olson (FMO) protein in Chlorobaculum tepidum. C. tepidum is a member of the Chlorobium bacteria and are closely related, yet distinct from, the Bacteroides phylum (2).
Characteristics of Green Sulfur Bacteria
Independent of light, life on earth would not exist. Biological photosynthesis is among the most important reactions on planet earth that allows life to thrive. Green Sulfur Bacteria are photosynthetic microbes that are capable of thriving in ecological regions that have extremely low levels of solar radiation. It has been proposed that certain types of green sulfur bacteria use what is know as Geothermal Light to supplement their chemotrophic metabolism. The emission of photons from geothermal vents could have provided a selective advantage to chemotrophic ancestors of modern day green sulfur bacteria that utilized light sensing systems to perform phototaxis towards sulfur-rich geothermal vents (3). On Earth, the discovery of a photosynthetic species capable of using geothermal radiation, rather than solar sources, helps support the hypothesis that life exists on other planets devoid of star-light.
In green sulfur bacteria, the harvesting of light is in part carried out by the Fenna-Matthews-Olson protein (FMO). Based on the current literature, the FMO protein is thought to be unique to the green sulfur bacteria (4). The chlorobiaceae contain Fe-S type reaction centers that share an ancestor with oxygenic photosynthetic organisms that utilize photosystem I (4).
Introduction to the FMO protein
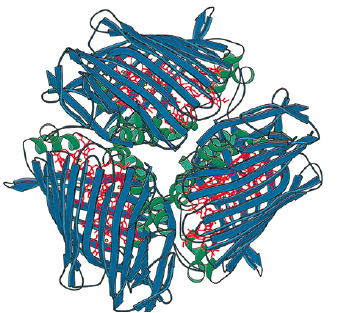
The Fenna-Matthews-Olson protein has been identified in all surveyed green sulfur bacteria to date (4). The FMO protein is also recognized by molecular biologists as the bacteriochlorophyll a protein (BChl a). The FMO (BChl a) protein was first described in the organism Prosthecochloris aestuarii (5, 6). BChl a is a hetero-trimeric protein that has been resolved at a maximum resolution of 1.9 angstroms (7). With this high resolution, knowledge of the structure has provided a background for the optical analysis of the protein. As is the case with many advancing work in science, the theoretical framework for this optical analysis has been disputed by other researchers (8). Determination
Chlorobaculum tepidum Fenna-Matthews-Olson (FMO) Antenna Protein
In C. tepidum, the Fenna-Matthews-Olson protein is part of a light-harvesting complex that consists of three major components: 1. Chlorosome 2. Aforementioned FMO protein, and 3. Reaction center. This system captures electromagnetic radiation from the sun and forms a funnel-like system that directs energy transfer (3). The role of the FMO protein is analogous to that of a wire connecting two electrodes. That is to say that the FMO protein provides the transfer of energy from the chlorosome to the reaction center.

Include some current research in each topic, with at least one figure showing data.
[edit] Section 2
Include some current research in each topic, with at least one figure showing data.
[edit] Discovering the Shape
Include some current research in each topic, with at least one figure showing data.
[edit] Conclusion
Overall paper length should be 3,000 words, with at least 3 figures.
[edit] References
(1)Washington University in St. Louis. "'Taco Shell' Protein: Orientation Of Antenna Protein In Photosynthetic Bacteria Described." ScienceDaily 9 April 2009. 12 April 2009 <http://www.sciencedaily.com¬ /releases/2009/04/090402171438.htm>.
(2)D.A. Bryant & N.-U. Frigaard (November 2006). "Prokaryotic photosynthesis and phototrophy illuminated". Trends Microbiol. 14 (11): 488. doi:10.1016/j.tim.2006.09.001
(3) Beatty, J.T.; Overmann, J.; Lince, M.T.; Mansket, A.K.; Lang, A.S.; Blankenship, R.E.; Van Dover, C.L.; Martinson, T.A.; Plumley, F.G. “ An obligately photosynthetic bacterial anaerobe from a deep-sea hydrothermal vent”. PNAS June 28, 2005 vol. 102 no. 26 9306-9310
(5) Olson, J. M. (1978). Bacteriochlorophyll a-proteins from green bacteria. In The Photosynthetic Bacteria (Clayton, R. K. & Sistrom, W. R., eds), pp. 161± 178, Plenum Press, New York
(6) Olson, J. M., Ke, B. & Thompson, K. H. (1976). Exciton interaction among chlorophyll molecules in bacteriochlorophyll a proteins and bacteriochlorophyll a reaction center complexes from green bacteria. Biochem. Biophys. Acta, 430, 524±537.
(7)Tronrud, D. E. & Matthews, B. W. (1993). Refinement of the structure of a water-soluble antenna complex from green photosynthetic bacteria by incorporation of the chemically determined amino acid sequence. In The Photosynthetic Reaction Center (Norris, J. & Deisenhofer, J., eds), vol. 1, pp. 13±21, Academic Press, New York.
(8)Gülen, D. (1996). Interpretation of the excited-state structure of the Fenna-Matthews-Olson pigment protein complex of Prosthecochloris aestuarii based on the simultaneous simulation of the 4K absorption, linear dichroism, and singlet-triplet absorption difference spectra: a possible excitonic explanation?. J. Phys. Chem. 100, 17683±17689.
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.