Chromohalobacter Salexigens
A Microbial Biorealm page on the genus Chromohalobacter Salexigens
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Chromohalobacter;
Species
C. Salexigens
Description and significance
This bacterium is a moderate halophile(salt loving), yet does not require high concentrations of sodium chloride. C. Salexigens is very flexible in that its salt requirements can be met by ions of other salts such as potassium, rubidium, ammonium, bromide, and others.
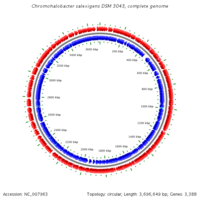
Genome structure
DNA Bases: 3696649
Chromosome Type: Circular
Total Genes: 3403
Protein Coding Genes: 3319 RNA Genes: 84 Pseudo Genes: 21
Cell structure and metabolism
The growth rate of Chromohalobacter salexigens DSM 3043 can be stimulated in media containing 0.3 M NaCl by a 0.7 M concentration of other salts of Na+, K+, Rb+, or NH4+, Cl-, Br-, NO3-, or SO4(2-) ions.
Edit: C. Salexigens is a moderate halophile that is capable of "making a living" in many various salt environments. In this way, it is a flexible bacterium. Placing C. Salexigens in media containing a 0.3M concentration of NaCL and a 0.7M concentration of (Na+, K+, Rb+, etc...) will stimulate its growth positively. [1]
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Chromohalobacter salexigens (bacterium) synthesizes and intracellularly accumulates molar concentrations of organic solutes, primarily ectoine and glycerol, respectively.
Edit: C. Salexigens produces and stores small amounts of ectoine and glycerol intracellularly. Small-scale production of these and other organic solutes is made possible via this organism. [2]
Interactions between C. Salexigens and other bacterium such as various strands of Salmonella allow for salinity tolerance modulation. In other words, this bacterium allows for other organisms to exist in environments they would otherwise not be able to cope with.
Pathology
Current research indicates that C. Salexigens is not known to be pathogenic.
Application to Biotechnology
The bacterium C. Salexigens synthesizes and accumulates compatible solutes in response to salt and temperature stress. The ectD gene, which is involved in the synthesis of the compatible solute hydroxyectoine, is essential for thermoprotection of the halophilic bacterium Chromohalobacter salexigens.
Edit: In response to salt and temperature stress C. Salexigens produces and stores various solutes. Some solutes, namely hydroxyectoine, are used in thermoregulation processes that protect C. Salexigens from extreme temperatures. [3]
Current Research
N(gamma)-acetyl-2,4-diaminobutyrate (NADA), the precursor of the compatible solute ectoine, was shown to function as an osmoprotectant for the non-halophilic bacterium Salmonella enterica serovar Typhimurium. The addition of NADA-containing extracts of an ectoine synthase mutant of the broad salt-growing halophile Chromohalobacter salexigens DSM 3043(T) could alleviate the inhibitory effects of high salinity in S. enterica, which lacks the ectoine biosynthetic pathway. NADA, purified from extracts of the mutant, protected S. enterica against salinity stress.
Edit: Research is being done on mutant C. Salexigens bacterium that synthesize ectoine. C. Salexigens mutants such as these can be used to produce N(gamma)-acetyl-2,4-diaminobutyrate (NADA). When said mutants are placed in conjunction with bacterium Salmonella Enterica Serovar Typhimurium, salinity stress typically present in this form of Salmonella ceased to persist. [4]
In another study, the long-term response of the broad-salt growing halophile Chromohalobacter salexigens DSM 3043T to salt stress has been investigated with respect to adaptive changes in membrane lipid composition. This study included the wild-type and three salt-sensitive, ectoine-deficient strains: CHR62 (ectA::Tn1732, unable to grow above 0.75 M NaCl), CHR63 (ectC::Tn1732, unable to grow above 1.5 M NaCl), and CHR64, which was able to grow in minimal medium M63 up to 2.5 M NaCl, but its growth was slower than the wild-type strain at salinities above 1.5 M NaCl. This mutant accumulated ectoine and hydroxyectoine as major compatible solutes, but also the ectoine precursor, N-gamma-acetyldiaminobutyric acid, and was found to be affected in the ectoine synthase gene ectC. The main phospholipids of the wild-type strain were phosphatidylethanolamine, phosphatidylglycerol (PG), and cardiolipin (CL). Major fatty acids were detected as 16:0, 18:1, and 16:1, including significant amounts of cyc-19:0, and cyc-17:0. CL and cyclopropane fatty acids (CFA) levels were elevated when the wild-type strain was grown at high salinity (2.5 M NaCl). Membranes of the most salt-sensitive trains CHR62 and CHR63, but not of the less salt-sensitive strain CHR64, contained lower levels of CL. The proportion of cyc-19:0 in CHR64 was three-fold (at 2.0M NaCl) and 2.5-fold (at 2.5 M NaCl) lower than that of the wild type, suggesting that this mutant has a limited capacity to incorporate CFA into phospholipids at high salt. The addition of 1 mM ectoine to cultures of the wild-type strain increased the ratio PG/CL from 1.8 to 3.3 at 0.75 M NaCl, and from 1 to 6.5 at 2.5 M NaCl, and led to a slight decrease in CFA content. Addition of 1 mM ectoine to the mutants restored the steady-state levels of CL and CFA found in the wild-type strain supplemented with ectoine. These findings suggest that exogenous ectoine might attenuate the osmostress response involving changes in membrane lipids.
Edit: In other research, C. Salexigens is being invested to better understand its long-term response to salinity stress regarding membrane modulation. Ectoine-deficient strains of C. Salexigens are unable to cope with salinity stress and undergo extensive membrane changes. The addition of ectoine to these deficient strains, however, allows these bacterium to maintain a salinity responsive membrane. [5]
References
Edited by Chris Wittrock, a student of Rachel Larsen and Kit Pogliano
