Clostridium cellulovorans: Difference between revisions
| (126 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | {{Uncurated}} | ||
==Classification== | ==Classification== | ||
[[File:Clostridiumtree.jpg|200px|thumb|right|Phylogenic tree]] | |||
• Kingdom - Bacteria <Br>• Phylum - Firmicutes <Br>• Class - Clostridia <Br>• Order - Clostridiales <Br> • Family - Clostridiaceae <Br> • Genus - Clostridium | |||
===Species=== | ===Species=== | ||
| Line 6: | Line 8: | ||
{| | {| | ||
| height="10" bgcolor="#FFDF95" | | | height="10" bgcolor="#FFDF95" | | ||
'''NCBI:[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi? | '''NCBI:[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Tree&id=1485&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy],[http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genome&Cmd=ShowDetailView&TermToSearch=26364 Genome]''' | ||
|} | |} | ||
Clostridium cellulovorans | ''Clostridium cellulovorans'' | ||
Other name: Clostridium cellulovorans strain 743B | Other name: ''Clostridium cellulovorans'' strain 743B | ||
==Description and Significance== | ==Description and Significance== | ||
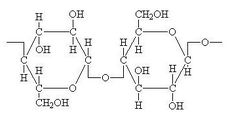
[[File:Cellulose1.jpg|230px|thumb|right|Cellulose]] | |||
<Br> Originally isolated from a batch methanogenic fermentation of hybrid poplar wood, ''Clostridium cellulovorans''(ATCC 35296) is rod shaped and non-motile. ''C. cellulovorans'' is an anaerobic, spore forming, gram-negative bacterium. ''C. cellulovorans'' is a mesophilic bacterium with optimum growth temperature of 37°C, though it can grow in a temperature range of 20 to 40°C. Optimum pH is 7.0, and the pH range of growth is 6.4 to 7.8. This organism produces extracellular enzyme complex known as cellulosomes which can degrade plant cell walls, notably cellulose. As most abundantly available potential source of fermentable sugars in the world are the cell walls in higher plants, utilization of such a vast resource for energy production would reduce the dependency on non-renewable fossil fuels. Hence, ''C. cellulovorans'' have potential industrial application for energy production. <Br> <Br> | |||
==Genome Structure== | ==Genome Structure== | ||
[[File:Cellulochromosome.jpg|190px|thumb|left|Chromosome map]] | |||
<Br> <Br> | |||
Genome sequencing of ''C. cellulovorans'' has been completed. ''C. Cellulovorans'' contains a circular chromosome with a length of 5,262,222 base pairs which is about 1 Mbp larger than the genomes from other cellulosomal clostridia. 31% of the genome is GC and 69% is AT. 57 cellulosomal genes were reported in ''C. cellulovorans''. ''C. cellulovorans'' contains large number of genes encoding non-cellulosomal enzymes which are more associated with polysaccharide (such as hemicelluloses and pectins) degradations other than cellulose. Scientists have found two novel genes encoding scaffolding proteins in ''C. cellulovorans'' genome. <Br> <Br> <Br> <Br> <Br><Br> <Br> | |||
==Cell Structure, Metabolism and Life Cycle== | ==Cell Structure, Metabolism and Life Cycle== | ||
''C. cellulovorans'' do not reduce sulfate and are obligate anaerobes. Cells are 0.7 to 0.9 by 2.5 to 3.5 µm in size and are non-motile rods, though peritrichous flagella were detected under electron microscopy. Both spores and vegetative colonies of ''C. cellulovorans'' are irregular, containing opaque edges and hollow centers. Spores are oblong occuring either centrally or subterminally within the mature sporangium. | |||
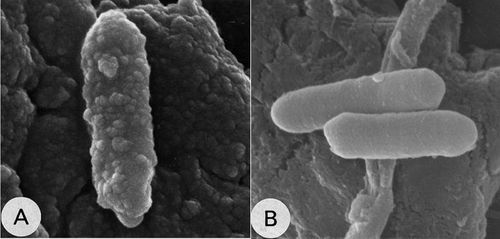
[[File:Final.jpg|border|thumbnail|500px|center|upright=0.75|''C. cellulovorans'' grown in cellulose (A) and in other medium (B)]] | |||
''C. cellulovorans'' obtains energy through the fermentation of cellulose and other carbohydrates. Apart from cellulose, ''C. cellulovorans'' ferments various carbon sources, such as xylan, pectin, cellobiose, glucose, fructose, galactose, sucrose, lactose and mannose and the fermentation products are hydrogen, carbon dioxide, acetate, butyrate, formate and lactate. When grown in cellulose, ''C. cellulovorans'' forms ultrastructural protuberances, which may be aggregation of smaller cellulosome complexes, also known as polycellulosomes. These protuberances were detected only in cellulose-grown cells and disappeared rapidly when other soluble carbohydrates were added to the growth medium. Cellulosomal components synergistically interact to catalyze the degradation of cellulose and hence, cellulosome acts as a macromolecular machine. <Br> | |||
==Ecology and Pathogenesis== | ==Ecology and Pathogenesis== | ||
[[File:Cellulosome.jpg|300px|thumb|right|Cellulosome]] | |||
''C. cellulovorans'' has no reported demonstration of mutualism in its environment, however, it does exhibit synergistic enzymatic relations within the cellulosome. Cellulosomes are complexes of cellulytic enzymes created to fuel the cell. The components of the cellulosome include dockerins, cohesions, enzymatic subunits, an anchoring protein, a carbohydrate binding domain and a scaffoldin subunit. Synergistic relations were discovered between cellulases (enzyme subunits), which affect cell wall degradation activity. Studies show individual cellulases liberate sugar at low rates but when xylanase is added to the cellulosome, rates increased dramatically. Synergistic relations between cellulosomal enzymes are important for studying effective degradation of organic plant life into usable fuel sources. Strong binding affinities of carbohydrate binding domains (CBD) in cellulosomes have also have provided important medical relevancy. The CBD can be used to bind cell signals for T cell activation which reduces the opportunity for graft-versus host disease in transplant patients. <Br><Br> ''Clostridium cellulovorans'' is non pathogenic to human beings. | |||
==References== | ==References== | ||
[http://www.ncbi.nlm.nih.gov/ | [http://www.ncbi.nlm.nih.gov/pubmed/10049808 Bayer, E. A., Shimon, L. J. W., Shoham, Y. and Lamed, R. "Cellulosomes - Structure and Ultrastructure". ''Journal of Structural Biology''. 1998. Volume 124. p. 221-234.]<Br> | ||
[http://www.ncbi.nlm.nih.gov/pubmed/10408097 Blair, B. G. and Anderson, K. L. "Regulation of Cellulose-Inducible Structures of ''Clostridium cellulovorans''. ''Canadian Journal of Microbiology''. 1999. Volume 45. p. 242-249.]<Br> | |||
[http://www.ncbi.nlm.nih.gov/pubmed/10449322/ Himmel, M. E., Ruth, M. F. and Wyman, C. E. "Cellulase for Commodity Products from Cellulosic Biomass". ''Current Opinion in Biotechnology''. 1999. Volume 10. p. 358-364.]<Br> | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2726447/ Maki, M., Leung, K. T., Qin, W. "The Prospects of Cellulase-Producing Bacteria for the Bioconversion of Lignocellulosic Biomass". ''International Journal of Biological Sciences''. 2009. Volume 5. p. 500-516.] <Br> | |||
[http://jb.asm.org/cgi/content/abstract/185/5/1518 Murashim, K., Kosugi, A. and Doi, R. H. "Synergistic Effects of Cellulosomal Xylanase and Cellulases from ''Clostridium cellulovorans'' on Plant Cell Wall Degradation". ''Journal of Bacteriology''. 2003. Volume 185. p. 1518-1524.] <Br> | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC240319/ Sleat, R., Mah, R. A. and Robinson, R. "Isolation and Characterization of an Anaerobic, Cellulolytic Bacterium, ''Clostridium-cellulovorans'' Sp-Nov". ''Applied Environmental Microbiology''.1984. Volume 48. p. 88-93.]<Br> | |||
[http://www.ncbi.nlm.nih.gov/pubmed/19948806 Tamaru, Y. et al. "Genome Sequence of the Cellulosome-Producing Mesophilic Organism ''Clostridium cellulovorans'' 743B". 2010. ''Journal of Bacteriology''. 2010. Volume 192. p. 901-902.]<Br> | |||
[http://www.ncbi.nlm.nih.gov/pubmed/20662379 Tamaru, Y., Miyake, H., Kuroda, K., Ueda, M. and Doi, R. H. "Comparative Genomics of the Mesophilic Cellulosome-Producing ''Clostridium cellulovorans'' and Its Application to Biofuel Production via Consolidated Bioprocessing". ''Environtal Technology''. 2010. Volume 31. p. 889-903.] <Br> | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC31190/ Tamaru, Y. and Doi, R. H. "Pectate lyase A, an enzymatic subunit of the ''Clostridium cellulovorans'' cellulosome". ''Proceedings of the National Academy of Sciences of the USA''.2001. Volume 98. p. 4125-4129.]<Br> | |||
==Author== | ==Author== | ||
Latest revision as of 19:08, 24 April 2011
Classification
• Kingdom - Bacteria
• Phylum - Firmicutes
• Class - Clostridia
• Order - Clostridiales
• Family - Clostridiaceae
• Genus - Clostridium
Species
Clostridium cellulovorans
Other name: Clostridium cellulovorans strain 743B
Description and Significance
Originally isolated from a batch methanogenic fermentation of hybrid poplar wood, Clostridium cellulovorans(ATCC 35296) is rod shaped and non-motile. C. cellulovorans is an anaerobic, spore forming, gram-negative bacterium. C. cellulovorans is a mesophilic bacterium with optimum growth temperature of 37°C, though it can grow in a temperature range of 20 to 40°C. Optimum pH is 7.0, and the pH range of growth is 6.4 to 7.8. This organism produces extracellular enzyme complex known as cellulosomes which can degrade plant cell walls, notably cellulose. As most abundantly available potential source of fermentable sugars in the world are the cell walls in higher plants, utilization of such a vast resource for energy production would reduce the dependency on non-renewable fossil fuels. Hence, C. cellulovorans have potential industrial application for energy production.
Genome Structure
Genome sequencing of C. cellulovorans has been completed. C. Cellulovorans contains a circular chromosome with a length of 5,262,222 base pairs which is about 1 Mbp larger than the genomes from other cellulosomal clostridia. 31% of the genome is GC and 69% is AT. 57 cellulosomal genes were reported in C. cellulovorans. C. cellulovorans contains large number of genes encoding non-cellulosomal enzymes which are more associated with polysaccharide (such as hemicelluloses and pectins) degradations other than cellulose. Scientists have found two novel genes encoding scaffolding proteins in C. cellulovorans genome.
Cell Structure, Metabolism and Life Cycle
C. cellulovorans do not reduce sulfate and are obligate anaerobes. Cells are 0.7 to 0.9 by 2.5 to 3.5 µm in size and are non-motile rods, though peritrichous flagella were detected under electron microscopy. Both spores and vegetative colonies of C. cellulovorans are irregular, containing opaque edges and hollow centers. Spores are oblong occuring either centrally or subterminally within the mature sporangium.
C. cellulovorans obtains energy through the fermentation of cellulose and other carbohydrates. Apart from cellulose, C. cellulovorans ferments various carbon sources, such as xylan, pectin, cellobiose, glucose, fructose, galactose, sucrose, lactose and mannose and the fermentation products are hydrogen, carbon dioxide, acetate, butyrate, formate and lactate. When grown in cellulose, C. cellulovorans forms ultrastructural protuberances, which may be aggregation of smaller cellulosome complexes, also known as polycellulosomes. These protuberances were detected only in cellulose-grown cells and disappeared rapidly when other soluble carbohydrates were added to the growth medium. Cellulosomal components synergistically interact to catalyze the degradation of cellulose and hence, cellulosome acts as a macromolecular machine.
Ecology and Pathogenesis
C. cellulovorans has no reported demonstration of mutualism in its environment, however, it does exhibit synergistic enzymatic relations within the cellulosome. Cellulosomes are complexes of cellulytic enzymes created to fuel the cell. The components of the cellulosome include dockerins, cohesions, enzymatic subunits, an anchoring protein, a carbohydrate binding domain and a scaffoldin subunit. Synergistic relations were discovered between cellulases (enzyme subunits), which affect cell wall degradation activity. Studies show individual cellulases liberate sugar at low rates but when xylanase is added to the cellulosome, rates increased dramatically. Synergistic relations between cellulosomal enzymes are important for studying effective degradation of organic plant life into usable fuel sources. Strong binding affinities of carbohydrate binding domains (CBD) in cellulosomes have also have provided important medical relevancy. The CBD can be used to bind cell signals for T cell activation which reduces the opportunity for graft-versus host disease in transplant patients.
Clostridium cellulovorans is non pathogenic to human beings.
References
Author
Page authored by Umesh Adhikari and Joe Araiz, student of Prof. Jay Lennon at Michigan State University.