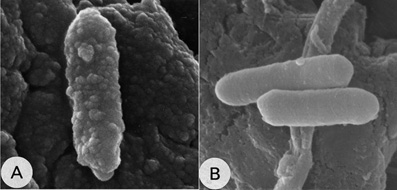
Clostridium cellulovorans
Classification
• Kingdom - Bacteria
• Phylum - Firmicutes
• Class - Clostridia
• Order - Clostridiales
• Family - Clostridiaceae
• Genus - Clostridium
Species
Clostridium cellulovorans
Other name: Clostridium cellulovorans strain 743B
Description and Significance
C. trachomatis is an obligate, aerobic, intracellular parasite of eukaryotic cells. It is a Gram-negative bacteria and has a cocci or rod shape. It has a cytoplasmic membrane and outer membrane like to Gram-negative bacteria but, it deficiencies of a peptidoglycan cell wall. C. trachomatis require growing cells in order to stay possible since it cannot create its own ATP. If there is no host organism, C. trachomatis cannot survive on alone [3]. C. trachomatis is the major cause of all sexually transmitted disease in the world; in the United States, its self, more than 4 million cases are diagnosed each year. It is also the chief cause of preventable blindness (affected by a chlamydia infection called trachoma) in the world [5]. C. trachomatis is one of the main causes of pelvic inflammatory disease (PID) and infertility in women [3]. It is very significant to understand and sequence the genome of C. trachomatis because it would aid us in better understanding its functions as a pathogen; the assets that allow it to survive within its human host and its virulence and biological capabilities as a pathogen. Therefore, an effort has been put forth to sequence a lot of the C. trachomatis genome [2].
Genome Structure
Describe the size and content of the genome. How many chromosomes? Circular or linear? Other interesting features? What is known about its sequence?
Genome sequencing of C. cellulovorans has been completed. C. Cellulovorans contains circular chromosomes, which length is 5,262,222 bp with GC content of 31%. Genome length of C. cellulovorans is about 1 Mbp larger than the genomes from other cellulosomal clostridia; 57 cellulosomal genes were reported in C. Cellulovorans 3. Number of predicted genes was the largest in C. cellulovorans as compared to other cellulosomal clostridia. C. Cellulovorans contains large number of genes encoding non-cellulosomal enzymes which are more associated with polysaccharides (such as hemicelluloses and pectins) degradation than to cellulose4.
Cell Structure, Metabolism and Life Cycle
Interesting features of cell structure; how it gains energy; what important molecules it produces.
C. Cellulovorans are 0.7 to 0.9 by 2.5 to 3.5 µm in size and are non-motile rods, though peritrichous flagella were detected under electron microscopy. Both spores and vegetative colonies of C. Cellulovorans are irregular containing opaque edge and a center devoid. Spores are oblong that occur either centrally or subterminally within the mature eporangium (1).
It produces plant cell wall degrading extracellular multienzyme complex called cellulosome5. Cellulosomal components synergistically interact to catalyze the degradation of cellulose and hence, cellulosome acts as a macromolecular machine 6. Cellulosome’s molecular weight is about 1 million and is able to hydrolyze crystalline cellulose7.Apart from cellulose, it ferments various carbon sources, such as xylan, pectin, cellobiose, glucose, fructose, galactose, sucrose, lactose and mannose to produce hydrogen, carbon dioxide, acetate, butyrate, formate and lactate 1.
Ecology and Pathogenesis
Habitat; symbiosis; biogeochemical significance; contributions to environment.
If relevant, how does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Clostridium Cellulovorans is non pathogenic to human beings.
References
(1) Sleat, R., Mah, R. A. & Robinson, R. Isolation and Characterization of an Anaerobic, Cellulolytic Bacterium, Clostridium-Cellulovorans Sp-Nov. Appl Environ Microb 48, 88-93 (1984).
2 Himmel, M. E., Ruth, M. F. & Wyman, C. E. Cellulase for commodity products from cellulosic biomass. Curr Opin Biotech 10, 358-364 (1999).
3 Tamaru, Y. et al. Genome Sequence of the Cellulosome-Producing Mesophilic Organism Clostridium cellulovorans 743B. J Bacteriol 192, 901-902 (2010).
4 Tamaru, Y., Miyake, H., Kuroda, K., Ueda, M. & Doi, R. H. Comparative genomics of the mesophilic cellulosome-producing Clostridium cellulovorans and its application to biofuel production via consolidated bioprocessing. Environ Technol 31, 889-903 (2010).
5 Tamaru, Y. & Doi, P. H. Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome. P Natl Acad Sci USA 98, 4125-4129 (2001).
6 Bayer, E. A., Shimon, L. J. W., Shoham, Y. & Lamed, R. Cellulosomes - Structure and ultrastructure. J Struct Biol 124, 221-234 (1998).
7 Doi, R. H. & Tamaru, Y. The Clostridium cellulovorans cellulosome: An enzyme complex with plant cell wall degrading activity. Chem Rec 1, 24-32 (2001).
Author
Page authored by Umesh Adhikari and Joe Araiz, student of Prof. Jay Lennon at Michigan State University.