Commercial applications of Rhodopseudomonas palustris
Introduction
Rhodopseudomonas palustris is an Alphaproteobacteria under the genus Rhodopseudomonas. R. palustris is a non-sulfur purple bacterium that is gram-negative with a rod shape. Cells are motile and the organism reproduces by means of budding. R. palustris is found in both aerobic and anaerobic environments including a wide variety of marine and soil ecosystems; coastal marine sediment and waste water treatment facilities for example[1]. Typically non-sulfur purple bacteria are phototrophic, however R. palustris has the capability to switch between different forms of metabolism depending on environmental conditions: photoautotrophy, photoheterotrophy, chemoautotrophy, and chemoheterotrophy.
Rhodopseudomonas palustris is the focus of extensive research because it has a multitude of potential metabolic processes. Research focusing on bioremediation and removal of waste is interested in R. palustris for its potential to break down aromatic compounds and waste in polluted environments. Rhodopseudomonas also has the capability to produce hydrogen as a product of nitrogen fixation, which researchers are looking into as a form of alternate energy produced from biofuel. R. palustris has also displayed the capability to generate electricity further increasing the scope of research surrounding the microbe. Climate change induced by global warming and pollution of environments are some of the largest environmental problems facing the modern world. R. palustris is interesting in that it could hold the potential to solving not just one, but both of these serious environmental issues. The extensive research on this microbe presents a possibility for unique commercial applications of the microbe as a source of alternate energy and in waste management and removal.
Metabolism
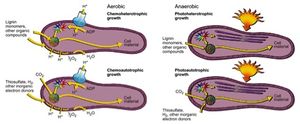
R. palustris has the capability to metabolize under aerobic and anaerobic conditions, meaning the organism can produce energy with or without oxygen present. Anaerobically, R. palustris can utilize photoheterotrophy and photoautotrophy, or photosynthesis[2] . The cell has the capability to fix carbon dioxide as biomass through photosynthesis, similar to green plants, but also R. palustris has the capability to obtain energy from light and other organic carbon forms via photoheterotrophy[3] . Under photoheterotrophic conditions, R. palustris utilizes organic compounds, most notably lignin monomers as a source of carbon[4] . When the cell is in the presence of conditions conducive to phototrophy, lamellar intracytoplasmic membranes hold bacteriochlorophylls and carotenoids, the photosynthetic pigments of the cell that make up the light harvesting complex, next to the cell membrane, which allows sunlight to be absorbed by the photosynthetic pigments[5] . The light energy absorbed is used to move electrons eventually resulting in the production of adenosine triphosphate (ATP). ATP is used as energy within the cell while the fixation of carbon dioxide or other organic compounds allows for the production of biomass and new cell material. Research has found that, R. palustris is able to respond to varying levels of light in its environment; in environments with lower light intensity the cell increases the amount of light harvesting complexes present to more effectively capture light energy[6] . Like photosynthetic plants, R. palustris possesses genes, which encode for RuBisCo, the enzyme responsible for the fixation of carbon dioxide in plants[7] . RuBisCo is used for the same fixation function in R. palustris photosynthesis, however unlike in plant photosynthesis oxygen is not a product of photosynthesis, as water is not split in the reaction and R. palustris utilizes thiosulfate, hydrogen and other inorganic electron donors[8] .
Research has shown that R. palustris has genes that resemble genes found in oxygenic photosynthetic bacteria that regulate a form of circadian clock for the organism[9] . While photosynthesis generates large amounts of energy for the cell during light hours, the energy produced would be limited during dark hours. The presence of these circadian clock genes would suggest that there is circadian regulation of reactions, in the cell notably for the process of nitrogen fixation[10] . Rhodopseudomonas is an example of a microbe that performs anoxic photosynthesis in which oxygen is not generated and instead nitrogen gas is fixed. R. palustris possesses genes that encode nitrogenase enzymes, which allows the organisms to take in atmospheric nitrogen gas and fix the nitrogen into ammonia and other fixed nitrogen forms.
Biodegredation
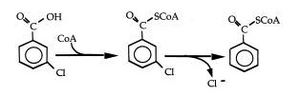
Include some current research, with at least one figure showing data.
Electricity Generation
Include some current research, with at least one figure showing data.
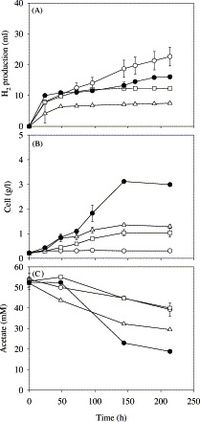
Hydrogen Production
Conclusion
References
- ↑ Larimer F. et al. “Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris”. Nature Biotechnology. 2004. Volume 22, p.55-61
- ↑ Larimer F. et al. “Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris”. Nature Biotechnology. 2004. Volume 22, p.55-61
- ↑ Larimer F. et al. “Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris”. Nature Biotechnology. 2004. Volume 22, p.55-61
- ↑ Larimer F. et al. “Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris”. Nature Biotechnology. 2004. Volume 22, p.55-61
- ↑ Mehrabi,S. et al." Identification and characterization of Rhodopseudomonas spp., a purple, non-sulfur bacterium from microbial mats"."Biomolecular Engineering". 2001.Volume 18,p.49-56.
- ↑ Larimer F. et al. “Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris”. Nature Biotechnology. 2004. Volume 22, p.55-61
- ↑ Scheuring, S. “The Photosynthetic Apparatus of Rhodopseudomonas palustris: Structures and Organization”. Journal of Molecular Biology. 2006.Volume 358, Issue 1, p.83-96.
- ↑ Scheuring, S. “The Photosynthetic Apparatus of Rhodopseudomonas palustris: Structures and Organization”. Journal of Molecular Biology. 2006.Volume 358, Issue 1, p.83-96.
- ↑ Scheuring, S. “The Photosynthetic Apparatus of Rhodopseudomonas palustris: Structures and Organization”. Journal of Molecular Biology. 2006.Volume 358, Issue 1, p.83-96.
- ↑ Scheuring, S. “The Photosynthetic Apparatus of Rhodopseudomonas palustris: Structures and Organization”. Journal of Molecular Biology. 2006.Volume 358, Issue 1, p.83-96.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2016, Kenyon College.
