Computer Logic in Microbial Systems
Introduction

By Jeremy Moore
Synthetic biology is a quickly evolving field that combines the sciences of molecular biology and biochemistry with engineering. One major task for synthetic biologists is harnessing cell’s innate ability to perform tightly concerted metabolic processes in order to produce molecules of industrial and medical relevance. In other words, one goal of synthetic biology is to create a programming language for cellular processes that can be altered with great precision to generate microbes with specific functions. Applying computer logic to living systems is challenging, as gene regulation is highly sensitive to the environment and requires a tightly controlled balance of regulatory factors[2]. Nonetheless, several methods have been developed to translate logical operators into genetic circuits.
Programmable cells have myriad applications to industry and medicine. As an example, a strain of Yersinia pseudotuberculosis was modified to invade cancerous cells in response to environmental conditions[1]. In this study, Y. pseudotuberculosis was equipped with synthetic genetic circuits that allowed it to detect changes in several environmental factors. In the presence of certain environmental conditions (such as high cell density and hypoxia), the strain expressed a protein that allowed it to invade mammalian cells (Figure 1). Applications like this could be used to create synthetic organisms capable of accomplishing highly specific tasks such as targeting specific tissues or compounds. Here, we will discuss several advancements in synthetic biology techniques used to program cells. By translating logical operations to genetic circuits, it is theoretically possible to design a cell to target a specific tissue or pathogen for drug delivery[2].
Logic Gates in Biological Context


The central principle to logic gate construction in living systems, is linking an output, such as expression of a fluorophore, to the expression of other genes in a regulatory genetic circuit[2]. For example, to get an OR logic gate (where one or both of two stimuli can activate the outpus), one could construct a system where two different promoters both activate a gene. Similarly, a NOR gate (where either response prevents a response) could be constructed as in Figure 2a, where expression of a repressor is controlled by two adjacent promoters. This repressor then prevents an output[2]. Gates that use AND logic can also be constructed. One method of AND gate construction is to have an output activated by a protein that requires a chaperone[2]. Thus, both the activator and chaperone, which must each have their own unique inducible promoter, are required to induce expression of the output.
The left side of Figure 2 shows several examples of possible logic gate designs[2]. In these examples, the output is some fluorophore, the expression of which is dependent on some interaction between two other promoters and the genes they serve. The right side of Figure 2 displays predicted expression of the fluorophore given expression of the two promoters that build the logical operation. Every logical operator can be constructed in several ways. A NOR gate, for example, can be built by placing two different promoters in front of a repressor gene. However, this gate, as shown in Figure 2b, can also be constructed by the use of invertible DNA elements containing an invertase gene. Invertases change the orientation of a particular DNA sequence from facing down to upstream of the gene it belongs to[4]. A NOR gate can be constructed with invertases if activation of either invertase flips a section of the promoter that activates an output. An AND gate can also be constructed using invertases; in this scenario, two invertases are designed to actually correct inversions in a promoter region. Activation of both promoters corrects both inversions, leading to transcription of the output gene.
The logic gate designs described above are not the only possible methods for achieving controlled expression of a gene. Throughout this article, several strategies for achieving programmable cells will be discussed. The major challenge with designing logic gates in bacteria is managing the noise of any particular signal. While any logical operation is theoretically possible in bacteria, certain logic gates are more efficient than others. For example, in Figure 3, an OR and NOR gate are compared. For each heat map, different pairs of promoters were used to demonstrate that not every pair behaves quite the same. Additionally, it was shown that OR gates are more susceptible to leaking (some intermediate states between on and off) than NOR gates which have nearly digital signals (few if any intermediate states between on and off) [3]. Because logical operators in bacteria rely on inherently stochastic interactions between regulator proteins and the promoters they bind, the strength of the promoter and its respective regulators determines how cleanly each gate performs.
Counting and Memory Function Using Invertases

Counters are important components of digital circuits and computer programs, so translating their function into living systems is necessary before implementing more complex computational functions. Counters exist in several natural forms. Telomere length, for example, is regulated by a counter of sorts. In the yeast, Saccharomyces cerevisiae, telomerase lengthening is initiated by the loss of a binding site for the Rap1p protein. This allows the cell to effectively count how many times it has divided in order to only initiate telomere elongation when absolutely necessary[5].
While yeast’s method of telomere length regulation was solved in the late 1990’s, uses for such counting logic have only recently been devised. A creative method of counting the number of times a particular event has occurred was described by Friedland et al. As shown in Figure 4, creating a line of several invertible DNA elements next to each other allows for the construction of genetic circuits with nearly digital levels of regulation[4]. In each element is an invertase enzyme that inverts the elements upon activation of an upstream promoter. This inversion then places a promoter in front of the next element in the sequence. The new promoter, upon introduction of its respective stimulus, either activates the next invertase in the sequences (thereby introducing a promoter at the next site), or, if it is on the final invertible sequence, activates the output. In the presented example, the output is activated after three independent arabinose pulses have been counted[4]. If more complex counting systems are necessary, the promoters on each invertible sequence can be altered. A functionality similar to that of a password is possible if promoters activated by different pulses are placed in each invertible sequence. For example, the first invertase could be arabinose inducible, but the promoter it places next could be IPTG inducible. Thus, in a two-invertase system, the output would only be activated by an arabinose pulse, followed by an IPTG pulse and not the reverse input [4].
The ability to count the number of times a particular event has occurred allows you to decide exactly when a particular response occurs. One could, for example, tie the inversion events to cell division[2]. Thus, the number of adjacent invertible sequences would decide exactly how many times a particular strain can divide before terminating. In medical applications of genetic circuit design, this alleviates some of the safety concern associated with using bacteria to treat infections[4]. One could theoretically design a bacteria that attacks a particular pathogen, but after a predetermined number of divisions, terminates itself.
Construction of Complex Logic


Oscillating Synthetic Gene Networks
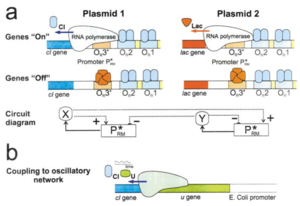
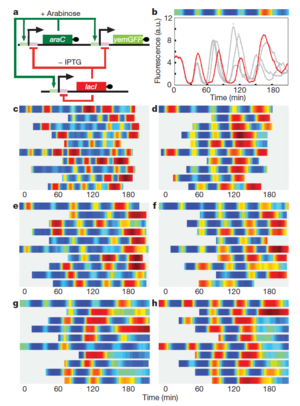
Circuit Interactions with the Host Cell
Conclusion
References
- ↑ 1.0 1.1 JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally Controlled Invasion of Cancer Cells by Engineered Bacteria. (2006). JMB 355: 619 – 627. doi:10.1016/j.jmp.2005.10.076.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Brophy JAN, Voigt CA. Principles of Genetic Circuit Design. (2014). Nature Methods 11(5): 508 – 520. DOI:10.1038/NMETH.2926.
- ↑ 3.0 3.1 3.2 3.3 http://www.nature.com/nature/journal/v469/n7329/abs/nature09565.html Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. (2011) Nature 469: 212 – 215. doi:10.1038/nature09565.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 . Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic Gene Networks that Count. (2009). Science 324(5931): 1199 – 1202.
- ↑ Marcand S, Gilson E. A protein-counting mechanism for telomere length regulation in yeast. (1997). Science 275: 986 – 990.
- ↑ https://journals.aps.org/prl/abstract/10.1103/PhysRevLett.88.148101 Hasty J, Dolnik M, Rottschafer V, Collins JJ. Synthetic Gene Network for Entraining and Amplifying Cellular Oscillations. 2002. Physical Review Letters 88(14) 148101.
- ↑ http://www.nature.com/nature/journal/v456/n7221/full/nature07389.html Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. 2008. Nature 456(27) 07389.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2017, Kenyon College.
