Corynebacterium diphtheriae: Difference between revisions
| Line 38: | Line 38: | ||
The genome contains what are termed Pathogenicity Islands (PAIs). These PAIs, of which C. diphtheriae has 13, contain local genetic anomalies in nucleotide composition or frequency, and many are closely associated with patches that code for tRNA. These PAIs code for the majority of fimbrial and fimbria-related genes, and iron-uptake systems which produce siderophores and lantibiotics. The ability to acquire iron allows C. diphtheriae to produce the Diphtheria toxin, which gives the bacteria its pathogenicity. | The genome contains what are termed Pathogenicity Islands (PAIs). These PAIs, of which C. diphtheriae has 13, contain local genetic anomalies in nucleotide composition or frequency, and many are closely associated with patches that code for tRNA. These PAIs code for the majority of fimbrial and fimbria-related genes, and iron-uptake systems which produce siderophores and lantibiotics. The ability to acquire iron allows C. diphtheriae to produce the Diphtheria toxin, which gives the bacteria its pathogenicity. | ||
Various methods were used to sequence the genome. The original strain of C. diphtheriae was obtained from the pharyngeal membrane of a 72-year-old female in 1997. The organism was then cultured and the DNA chromosome extracted. The genome sequence was obtained from 60,750 end sequences (with 9.2x coverage) derived from two pUC18 genomic shotgun libraries. All repeats were bridged using read-pairs or PCR, after which the genome was assembled. Computer analysis was used to identify possible genes and coding segments. | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
Revision as of 14:12, 3 May 2007
A Microbial Biorealm page on the genus Corynebacterium diphtheriae
Classification
Eubacteria
Higher order taxa
Actinomycetales
Species
|
NCBI: Taxonomy |
Corynebacterium diphtheriae
Description and significance
- Picture from textbookofbacteria.net to be included hopefully
- Note - include picture of Emil Von Behring
C. diphtheriae is a gram-positive, aerobic, nonmotile, toxin-producing, rod-shaped bacteria belonging to the order Actinomycetales, which are typically found in soil, but also have pathogenic members such as streptomyces and mycobacteria. C. diphtheriae is best known for causing the disease Diphtheria in human begins, which results from production of Diphtheria toxin in conjunction with infection by a bacteriophage which provides it with the toxin-producing gene. Because historically it has been a very deadly disease, especially for children where mortality rates before vaccines and antitoxin amounted to nearly 80%, it has been heavily studied. Oddly enough, mice and other rodents are naturally immune to the Diphtheria toxin so it has been difficult to study Diphtheria in the lab.
Diseases resembling Diphtheria were described as early as the 4th Century BCE by Hippocrates. It was named in 1826 by French physician Pierre Bretonneau, who gave it the name Diphtheria from the Greek word which means "leather hide". It was named "leather hide" because of the leathery layer that grows on the tonsils, throat, and nasal passages. Corynebacterium diphtheriae was first described in 1884 by Edwin Klebs and Friedrich Loffler, giving it the name Klebs-Loffler bacillus by which it used to be known.
In 1888, Emile Roux discovered that a toxin produced by C. diphtheriae was the causative agent in the disease Diphtheria. Emil Von Behring (1854-1917) was one of the first pioneers in the race to fight Diphtheria. He studied C. diphtheriae from 1889 onwards, especially in animals. When he discovered that the serum of anthrax-resistant rats was able to kill the anthrax bacillus, he researched such possibilities for Diphtheria, eventually using the serum of Diphtheria toxin-resistant guinea pigs, and ultimately horses. Eventually he developed antiserum, based on his experiments, as well as theories developed by a previous colleague Shibasaburo Kitasato. By 1892, the antiserum was ready for commercial production. In 1891, Behring was awarded the Nobel Prize in Medicine for his work.
In 1923, the work of Alexander Glenny, Barbara Hopkins, and Gaston Ramon produced a vaccine for Diphtheria. Glenny and Hopkins found that formaldehyde could prevent the Diphtheria toxin from being toxic, while Ramon discovered that the nontoxic form still retained its antigenic qualities. The form of the toxin which results from denaturing with formaldehyde is called an anatoxin, "toxoids", or "formalin-toxoids", and is used in vaccinations worldwide.
- Will include a picture of Balto
Outbreaks of Diptheria were common earlier in the 18th and 19th century, but the introduction of the vaccine caused a temporary decline in the number of cases. One of the more well-known outbreaks of Diphtheria was commemorated in the animated film "Balto", a story about the dog sled team (whose lead dog was named Balto) which brought the Diphtheria antiserum from Nenana to Nome, which was geographically isolated during the epidemic. They traveled 700 miles in 6 days through temperatures of minus 40 degrees Celsius and blizzard-speed winds. The dog sled team saved hundreds of lives, the story was sensationalized worldwide, and a statue of Balto, the lead dog, was erected in New York's Central Park.
Despite having a vaccine and antiserum, there is currently an increase in the number of cases in the United States and many countries in eastern Europe. Research is being done to determine why new epidemics are being seen - whether it is a result of a mutation in C. diphtheriae, or if it is from environmental and social factors such as poor immunization rates and unsanitary conditions.
Genome structure
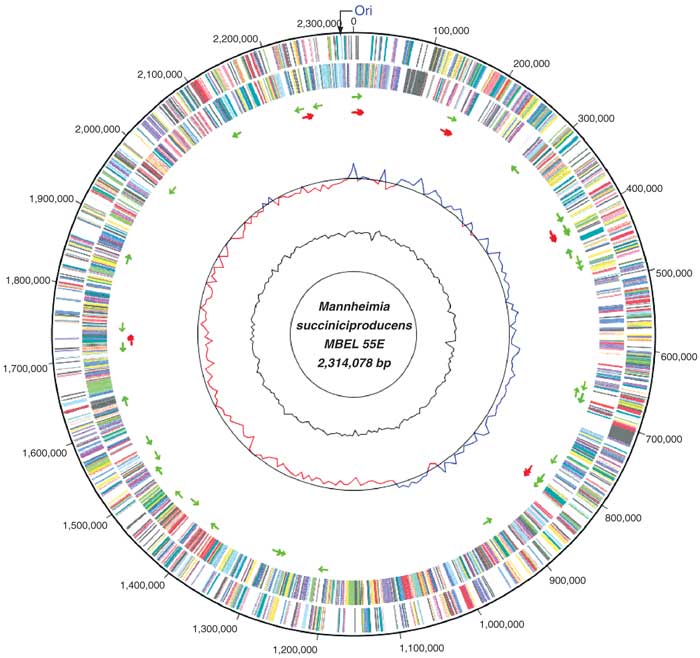
Image from Wishart Research Group, University of Alberta
The genome of C. diphtheriae contains 2,488,635 nucleotides, 87% of which are coding. This single circular chromosome contains 2,389 genes from which 2,272 proteins are coded. It contains no plasmids. There are 69 structural RNA, 15 coding for 5S, 23S, and 16S Ribosomal RNA, and 54 coding for the 20 amino acid tRNA molecules. The GC content is 53%, though it is not evenly distributed - it has higher proportion of GC pairs near the origin of the plasmid (around 55%), compared to the region near the terminus (49%)
The genome contains what are termed Pathogenicity Islands (PAIs). These PAIs, of which C. diphtheriae has 13, contain local genetic anomalies in nucleotide composition or frequency, and many are closely associated with patches that code for tRNA. These PAIs code for the majority of fimbrial and fimbria-related genes, and iron-uptake systems which produce siderophores and lantibiotics. The ability to acquire iron allows C. diphtheriae to produce the Diphtheria toxin, which gives the bacteria its pathogenicity.
Various methods were used to sequence the genome. The original strain of C. diphtheriae was obtained from the pharyngeal membrane of a 72-year-old female in 1997. The organism was then cultured and the DNA chromosome extracted. The genome sequence was obtained from 60,750 end sequences (with 9.2x coverage) derived from two pUC18 genomic shotgun libraries. All repeats were bridged using read-pairs or PCR, after which the genome was assembled. Computer analysis was used to identify possible genes and coding segments.
Cell structure and metabolism
- WORK IN PROGRESS*
Describe any interesting features and/or cell structures; how it gains energy; what important molecules it produces.
Cell Exterior
Being gram-positive, C. diphtheriae has a cell membrane and a lipid-rich murein layer outside the membrane. Like M. tuberculosis, another Actinomycete, C. diphtheriae has an arabinogalactan polymer which anchors a lipid rich domain to the murein layer. Corynomycolic acids, alpha-alkyl beta-hydroxy fatty acids, are produced through a Claisen-like condensation reaction between two fatty acyl chains.
Also in the exterior are the fimbriae (pili). Not much structural analysis has been done, however it is very likely that sortase-like proteins are produced to aid in anchoring the fimbriae and possibly polymerizing them. C. diphtheriae has six genes which code for putative sortases - five are localized on PAIs, none of which are found in closely related non-pathogenic corynebacteria (C. glutamicum and C. efficens). The absence of these genes in the non-pathogenic relatives suggests that those genes increase the pathogenicity of C. diphtheriae through the fimbriae, most likely by aiding the bacterium invade and adhere to host cells. The similarity to the sortase-related fimbrial system of Actinomyces, and the newness of the genes in the C. diphtheria genome, suggest that they were newly acquired, possibly the cause of the recent upsurge in outbreaks of Diphtheria.
Metabolic processes
Pathways discovered for metabolism include the complete pentose phosphate pathway, glycolysis, and gluconeogenesis, based on the enzymes present in the cell. The citric acid cycle is present, however C. diphtheriae lacks succinyl-CoA synthetase and uses an enzyme transcribed by a homologue of the Clostridium kluyveri cat1 gene, which acts as a coenzyme A transferase rather than a synthetase. De novo amino acid biosynthesis is also present in C. diphtheriae, as well as both anaerobic and aerobic respiration pathways, though C. diphtheriae is primarily aerobic. Production of purines is complete, however of the purines, the cytidine production pathway lacks the final cytidine triphosphate synthetase, which is present in other related organisms such as C. efficens and M. tuberculosis.
Specialized Iron Intake and Diphtheria Toxin
Diphtheria toxin is the most significant molecule produced by C. diphtheriae. The tox gene which codes for the Diphtheria toxin is located on the right-handed end of an integrated corynephage. C. diptheriae contains the protein DtxR, an iron-dependent negative-regulatory protein which prevents the tox gene from being transcribed under high iron conditions, as well as the high-energy iron uptake system of the bacterium. Because a host often reacts to infection by lowering iron levels, the bacteria have developed a mechanism which couples Diphtheria toxin gene expression with low iron levels.
To intake iron, C. diphtheriae has seven different systems, two of which have been identified. The siderophore system creates ferrisiderophores, molecules specialized in binding to iron and bringing it into the cell for usage.
Ecology
- WORK IN PROGRESS*
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
- WORK IN PROGRESS*
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Application to Biotechnology
- WORK IN PROGRESS*
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
- WORK IN PROGRESS*
Enter summaries of the most recent research here--at least three required
References
- WORK IN PROGRESS*
Edited by student Rachael Spradley of Rachel Larsen
