Cytophaga Flavobacterium
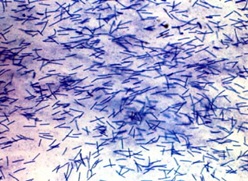
Members of the Cytophaga-Flavobacterium group are Gram- negative, heterotrophic,non-spore forming rods that possess gliding
motility (Kirchman. 2002). Bacteria in this group belong to the genus Flavobacterium, and the genus Cytophaga and
are characterized by their phenotype and 16S rRNA sequence (Kirchman. 2002).These bacteria can be distinguished by their
flexirubin type pigmentationproduced only by this group (Kirchman. 2002). Furthermore, members under this
group are mainly aerobic with some facultative anaerobes. (Reichenbach et al.,1991).
Due to several rearrangements of classification of organisms under the Cytophaga genus and Flavobacterium genus, researches
regularly unite these genera into a single group called the Cytophaga-Flavobacterium group, the Cytophaga-Flexibacter-Bacteriodes (Mayrberger. 2011),
or Bacteroidetes (Kreig. 2010). This broad phylogenetic label is used because of the limited understanding of specific species within each genus.
Environment
Bacteroidetes can be found globally in every biosphere (Kirchman. 2002). The group thrives in the rhizosphere which is an area of the soil that is directly associated with plant roots (Lines-kelly. 2005). In these soil aggregates, members influence the carbon cycle through the degradation of organic carbon compounds (Mayrberger. 2011). The Cytophaga-Flavobacterium group also prospers in the costal water, offshore water, sediments, hydrothermal vents, and the polar region (Alonso et al., 2007). Through performing FISH (fluorescence in situ hybridization) to localize specific rRNA targets and performing cell counts, Bacteroidetes are often found to be the most abundant group in the pelagic zone (Alonso et al., 2007). In these niches, the group can be found free living, attached to organic compounds and associated with marine plankton and animals (Alonso et al., 2007).
Genome Identification
CF319a is the most frequently used FISH oligonucleotide probe to examine the diverse rRNA sequences in Bacteroidetes (Kirchman. 2002). This probe can also hybridize some members of the Bacteroides-Prevotella group (Weller et al., 2000).
It is important to note that CF319a does not recognize some genera of Bacteroidetes such as Cytophaga hutchinsonii, and studies conducted using this probe underestimate the Cytophaga- Flavobacterium abundance (Kirchman. 2002). Weller et al. (2000) suggests that a habitat-specific-probe should be developed that include genera missed by CF319 as well as genera recognized by this probe, but currently no such probe has been developed. In total only 781 16S rRNA gene sequences of the Cytophaga-Flavobacterium group have been sequenced, and this represents only 5% of all 16 S rRNA sequences in this group (Kirchman. 2002).
Decomposition of Organic Matter
The Cytophaga-Flavobacterium group are chemoorganotrophs (Kirchman, D. 2002) that can degrade cell wall, cellulose, cellobiose, carboxymethylcellulose, chitin, starch, inulin, keratin, xylan, pectin, pectate, agarpectin, agar, laminarin, lipids, DNA, and RNA (Mayberger. 2011).
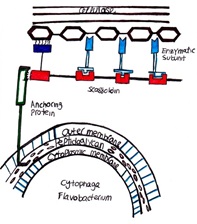
Some members of the Bacteroidetes possess the ability to degrade cellulose and glide over their substrate as they decompose and consume it (Thomas 2011). Since cellulose is too large to be transported into the cell, the bacteria secrete extracellular enzymes that assemble in the environment to form a complex called the cellulosome to degrade cellulose into products that can be imported into the cell (Bayer.1998) The cellulosome is tethered to the cell wall and contains enzymes such as exo-glucanases, endo-glucanses, xylanases, and hemicellulose. These enzymes are organized by the non-catalytic central protein scaffoldin (Bayer.1998). Scaffoldin is also responsible to substrate binding (Bayer.1998).
Involvement in the Carbon Cycle
Cytophaga- Flavobacterium group exacerbates the loss of organic carbon in the soil which is a causation of global climate change (Mayrberger. 2011). Bacteroidetes actively participate in the soil aggregate cycle by decomposing and recycling macroaggregates and releasing microaggregates and organic compounds to be incorporated into a new aggregate (Mayrberger. 2011). The loss of organic matter is due to the increased rate of aggregate cycling which cause carbon mineralization by Bacteroidetes to be faster than plant fixation of carbon dioxide (Mayrberger. 2011). Since organic matter in the soil provides nutrients for plants, the amplified loss of organic carbon from soil to the atmosphere leads to a lower agricultural yield and subsequently lowers the sequestration of carbon dioxide into the soil (Mayrberger. 2011). This is an example of a positive feedback response that leads to the increased concentration of carbon dioxide in the atmosphere and contributes to climate change.
Columnaris Disease
Flavobacterium columnare enters the fish through an opening such as an open wound to cause the Columnaris disease. The bacteria then proliferate and grow confluently on the surface of the gill producing protein degrading enzymes that result in cell death of gill filaments and eventually death of the fish (Durborrow. 1998). Infected fish often have yellowish –brown lesions on its gill and mouth. Flavobacterium columnar regularly inhibits marine environments and can cause the disease under regular conditions (Durborrow. 1998). Its growth is favourable when the fishes are under stressful conditions such as low oxygen, high ammonia, high water temperature, crowding, and injuries. In an in-vitro setting, the bacteria can be identified by its yellow pigmented rhizoid colonies, and its production of gelatin-degrading enzymes, and sulfate-degrading-enzymes (Durborrow. 1998).
References
1. Alonso, C., et al. "High local and global diversity of Flavobacteria in marine plankton." Environmental microbiology, 2007. Volume 9 Issue 5. p. 1253-1266. 2. Bayer, E.A., Shimon, L.J.W., Shoham, Y., and Lamed, R., “Cellulosomes—Structure and Ultrastructure” JOURNAL OF STRUCTURAL BIOLOGY . 1998, Volume124, p221–234, ARTICLE NO. SB984065. 3. Burmeister , Maren, and Heather Owen. “cover image.” American Society of Microbiology. 2007.Web, Retrieved 23 Nov 2012. http://www.google.ca/imgres?um=1&hl=en&tbo=d&biw=1241&bih=606&tbm=isch&tbnid=Fc0S7iRcEqMw3M:&imgrefurl=http://aem.asm.org/content/73/11.cover-expansion&docid=MkQZOFJwIsJazM&imgurl=http://aem.asm.org/content/73/11/F1.medium.gif&w=384&h=440&ei=cBGvUI2bBIf0iQKCoIGAAQ&zoom=1&iact=hc&dur=347&sig=100460146333430452513&page=1&tbnh=146&tbnw=139&start=0&ndsp=19&ved=1t:429,r:1,s:0,i:99&tx=116&ty=85&vpx=181&vpy=176&hovh=147&hovw=140 4. Durborow, R.M., Thune, R.L., Hawke, J.P., and Camus, A.C., “Columnaris Disease-A Bacterial Infection Caused by Flavobacterium columnare.” Southern Regional Aquaculture Center.1998, No. 479. http://www.google.ca/imgres?num=10&um=1&hl=en&tbo=d&biw=1241&bih=606&tbm=isch&tbnid=0E8lZsdR2VXqLM:&imgrefurl=http://www.soi.wide.ad.jp/class/20070046/slides/06/70.html&docid=lv7UMdrmfk3nZM&imgurl=http://www.soi.wide.ad.jp/class/20070046/slides/06/img/70.png&w=480&h=360&ei=iCGvUL_hJIfNigLUm4DwCA&zoom=1&iact=rc&dur=779&sig=100460146333430452513&sqi=2&page=1&tbnh=142&tbnw=213&start=0&ndsp=15&ved=1t:429,r:8,s:0,i:106&tx=96&ty=79 5. “Flavobacterium Power Chemicals.” Water Services. 2004. Web, Retrieved 23 Nov 2012. http://www.google.ca/imgres?um=1&hl=en&tbo=d&biw=1241&bih=606&tbm=isch&tbnid=D6WWE8rzZ1M1FM:&imgrefurl=http://www.power-chemicals.com/bio/flavobacterium.htm&docid=uegPVcUisV7DwM&imgurl=http://www.power-chemicals.com/bio/images/Gallery/Flavobacterium.jpg&w=385&h=281&ei=5BOvULbNLsvqiwLZwoFw&zoom=1&iact=hc&vpx=562&vpy=155&dur=1186&hovh=192&hovw=263&tx=199&ty=99&sig=100460146333430452513&page=1&tbnh=144&tbnw=198&start=0&ndsp=20&ved=1t:429,r:4,s:0,i:94 6. Kirchman, D.L. "The ecology of Cytophaga–Flavobacteria in aquatic environments." FEMS Microbiology Ecology, 2002. Volume 39 Issue 2. p. 91-100.
7. Krieg, N. R., Ludwig, W., Euze´by, J. & Whitman, W. B. “Phylum XIV. Bacteroidetes phyl. nov. Bergey’s Manual of Systematic Bacteriology. 2010, 2nd edn, vol. 4, p. 25. Edited by N. R. Krieg, J. T. Staley, D. R. Brown, B. P. Hedlund, B. J. Paster, N. L. Ward, W. Ludwig & W. B.
8. Lines-Kelly, R., “The rhizosphere: soil biology basics.” Profitable and sustainable primary industries. 2005. Web, Retrieved 23 Nov 2012, www.dpi.nsw.gov.au
9. Mayrberger, JM. "Studies of genera cytophaga-flavobacterium in context of the soil carbon cycle." Dissertation Abstracts International, 2011, Volume. 73, No. 01, Suppl. B. p. 1- 115
10. Reichenbach, H. “ The order Cytophagales.” In The Prokaryotes, 2nd edn, vol. 4, pp. 3631–3675. Edited by A. Balows, H. G. Trüper, M. Dworkin, W. Harder & K. H. Schleifer.
11. Takenaka, A., “Cellulosome model of C. thermocellum.” SOI Asia biotechnology special lecture. 2008. Web, Retrieved 23 Nov 2012.
12. Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. “Environmental and gut bacteroidetes: the food connection.” Front Microbiol.2011 Volume 2. p. 93.
13. Weller, R, Glöckner, F.O, Amann, R “16S rRNA-targeted oligonucleotide probes for the in situ detection of members of the phylum Cytophaga-Flavobacterium-Bacteroides.” Syst. Appl. Microbiol. , 2000. Volume 23. p. 107–114.