Denitrovibrio acetiphilus: Difference between revisions
No edit summary |
|||
| Line 1: | Line 1: | ||
{{Uncurated}} | {{Uncurated}} | ||
==Classification== | ==Classification== | ||
Revision as of 23:03, 11 April 2012
Classification
- THIS PAGE IS STILL IN THE CONSTRUCTION PHASE AND IS NOT COMPLETE**
Domain: Bacteria;
Phylum: Deferribacteres
Class: Deferribacteres
Order: Deferribacterales
Family: Deferribacteraceae
Genus: Denitrovibrio(1)
Species
Species: acetiphilus (2)
|
NCBI: Taxonomy |
Denitrovibrio acetiphilus
Description and Significance
Denitrovibrio acetiphilus is a marine, mesophilic, obligately anerobic organism. D. acetiphilus respires through nitrate reduction. [3]
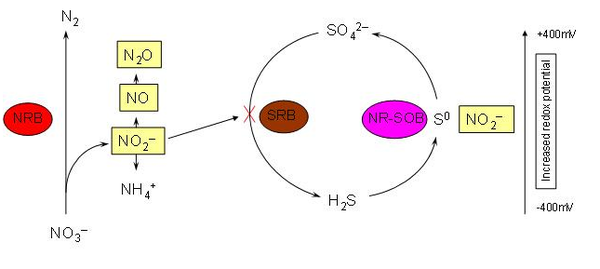
Denitrovibrio acetiphilus is of phylogenetic interest because there are currently only six genera described in the family Deferribacteraceae. The organism was found in a laboratory column reducing nitrate to ammonia. The column simulated off-shore oil drilling conditions, and the presence of this organism will prove beneficial to stopping the process of undesired hydrogen sulfide formation. When nitrate is added to the system with sulfate reducing populations, production is overcome by these nitrate reducing bateria. [3]
Genome Structure
Denitrovibrio acetiphilus has two known completed genome sequences, of the order Deferribacterales and the class Deferribacteres. This is the sole class in the phylum Deferribacteres. The 3,222,077 bp genome is comprised of one main circular chromosome. There is 3,085 total genes of which 3,034 are protein-coding genes and 51 are RNA genes. This research is part of the G enomic E ncyclopedia of B acteria and A rchaea project (GEBA) [3]. A 16S rRNA gene analysis shows the strain of N2460T D. acetiphilus belongs to the Bacteria, with the closest relatives being ‘Geovibrio ferrieducens’, ‘Deferribacter thermophilus’, and ‘Flexistipes sinusarabici’ [2] (See Figure 2).
Cell Structure, Metabolism and Life Cycle
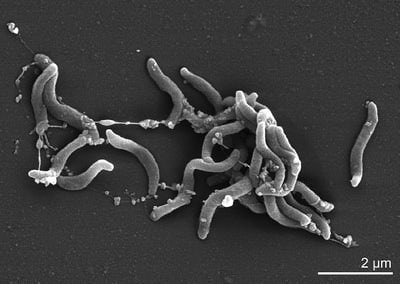
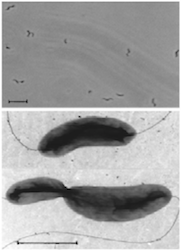
D. acetiphilus is a gram negative bacterium that possesses a curved rod shaped structure, similar to those in the genus, Vibrio. However, unlike those in the genus, D. acetiphilus are motile by bipolar flagellation and exhibit a rapid corkscrew movement. Cells of D. acetiphilus measure at 0.5-.07 by 1.7-2.0 µm and can be seen as singles, in pairs, or arranged in long chains. C16 and C18 fatty acids are major components of the cell membrane. [2]
Denitrovibrio acetiphilus(strain N2460T) reduces nitrate to ammonia by supplementation of acetate which acts as a substrate and electron donor. Nitrate therefore acts as an electron acceptor and acetate reduction to nitrite (an intermediate during reduction to ammonia) acts as the primary energy source. Further reduction of nitrite to ammonia is a possible electron source (electron sink) from which coenzymes are regenerated or may act as a detoxification system. Selective enrichment from an oil reservoir model column demonstrates the ability of this organism to grow at temperatures ranging from 4 to 40 degrees Celsius with optimal growth temperature between 35-37 degrees Celsius. Optimum growth also requires the presence of vitamins and a pH range of 6.5-8.6 in addition to supplementation of acetate to the medium. D.acetiphilus is unable to grow in the presence of oxygen and this organism is therefore labeled a strict anaerobic acetate specialist. Activity of 2-oxoglutarate dehydrogenase suggests acetate is oxidized via the citric acid cycle. Denitrovibrio acetiphilus (N2460T) is considered the nitrate reducing metabolic analogue of sulfate reducing Desulfobacter (also able to oxidize acetate through the citric acid cycle). From this information, D.acetiphilus can be described as a marine, mesophilic, obligate anaerobe that obtains its energy by dissimilatory nitrate reduction.
Ecology
As mentioned previously, this bacterium sheds insight into optimizing oil recovery in off-shore drilling. Presently, seawater is injected during oil recovery to maintain desired pressures and increase efficiency. In most cases the seawater contains high concentrations of sulfate which in turn results in a higher population of sulfate-reducing bacteria at these mine sites [3]. Through anaerobic respiration these sulfate-reducing bacteria produce hydrogen sulfide (H2S) that can potentially sour the oil reserves and lead to poor gas quality, corrosion and poor separation characteristics in further processing. Hydrogen sulfide may also harm the platform operators on oil rigs [4]. In many cases H2S inhibits the senses of the nose making it almost undetectable in the absence of air monitors. Experiments have shown that adding nitrate to these reservoirs may reduce the production of hydrogen sulfide by stimulating the growth of nitrate-reducing bacteria and inhibiting sulfate-reducing bacteria. This microbial shift may provide a more economical solution to minimizing H2S by eliminated the need for expensive biocides that are currently used to treat oil reserves.[3]
References
[1] Denitrovibrio acetiphilus taxonomy - (http://www.catalogueoflife.org/details/species/id/4257206)
[2] Myhr, S., and Torsvik, T. "Denitrovibrio acetiphilus, a novel genus and species of dissimilatory nitrate-reducing bacterium isolated from an oil reservoir model column". International Journal of Systematic and Evolutionary Microbiology". 2000. Volume 50. pgs. 1611–1619 (http://ijs.sgmjournals.org/content/50/4/1611.full.pdf).
[3] Kiss, H., Lang, E., Lapidus, A., Copeland, A., Nolan, M., Glavina Del Rio, T., Chen, F., Lucas, S., Tice, H., Cheng, J., Han, C., Goodwin, L., Pitluck, S., Liolios, K., Pati, A., Ivanova, N., Mavromatis, K., Chen, A., Palaniappan, K., Land, M., Hauser, L., Chang, Y., Jeffries, C., Detter, J., Brettin, T., Spring, S., Rohde, M., GöKer, M., Woyke, T., Bristow, J., Eisen, J., Markowitz, V., Hugenholtz, P., Kyrpides, N., Klenk, H.. "Complete genome sequence of Denitrovibrio acetiphilus type strain (N2460T)". Standards in Genomic Sciences, North America, 2, jun. 2010. Available at: (http://www.standardsingenomics.org/index.php/sigen/article/view/sigs.892105) . Date accessed: 21 Mar. 2012.
[4] "Nitrate: A Cost Efficient Alternative to Biocides." Norwegian Centre of Excellence. Web. 11 Apr. 2012. <http://www.cipr.uni.no/contentitem.aspx?ci=1546>.
Authors
Page authored by Rodney Tocco, Minh Tran, Samantha Wengert, and Eric Werner, students of Prof. Jay Lennon at Michigan State University.
<-- Do not remove this line-->