Dental Plaque Biofilms: Difference between revisions
| Line 50: | Line 50: | ||
[[Image:Figure1.jpg|250 x 150 px|right|thumb|(a) In the presence of <i>S. cristatus</i> CC5A <i>P. gingivalis</i> was inhibited from colonizing on the plaque biofilm. (b) In the presence of <i>S. gordonii</i> G9B, however, colonization of <i>P. gingivalis</i> occurred, thus indicating that <i>S. gordonii</i> was not involved in a cell signaling mechanism similar to <i>S. cristatus</i>. Red = Streptococcal cells, Green = <i>P. gingivalis</i>, and yellow = colonized bacteria. [http://jb.asm.org/cgi/reprint/182/24/7067<6>]]] | [[Image:Figure1.jpg|250 x 150 px|right|thumb|(a) In the presence of <i>S. cristatus</i> CC5A <i>P. gingivalis</i> was inhibited from colonizing on the plaque biofilm. (b) In the presence of <i>S. gordonii</i> G9B, however, colonization of <i>P. gingivalis</i> occurred, thus indicating that <i>S. gordonii</i> was not involved in a cell signaling mechanism similar to <i>S. cristatus</i>. Red = Streptococcal cells, Green = <i>P. gingivalis</i>, and yellow = colonized bacteria. [http://jb.asm.org/cgi/reprint/182/24/7067<6>]]] | ||
One such example of this involves the gram-negative, anaerobic bacteria <i>Porphyromonas gingivalis</i>. <i>P. gingivalis</i> is a predominant pathogen in severe cases of adult periodontitis and attaches to the tooth surface after the initial colonization of the gram-positive <i>Streptococcus</i> species. The colonization of <i>P. gingivalis</i> is key in the transformation of the bacterial biofilm from a commensal plaque to the pathogenic form observed in periodontitis [6]. Its colonization is also dependent on its attachment to oral surfaces, which is accomplished via a fimbriae-mediated adhesion. The <i>fimA</i> gene encodes for this subunit protein, which can be regulated environmentally [6]. Interestingly, the <i>P. gingivalis</i> <i>fimA</i> gene expression was found to be severely downregulated by the unrelated, gram-positive <i>Streptococcus cristatus</i> CC5A, which can be found in the initial colonization of dental biofilms. By interfering with the gene that encodes for the fimbriae-subunit protein, <i>S. cristatus</i> is able to prevent <i>P. gingivalis</i> fimbriae attachment to the biofilm, thus resulting in an easier removal of <i>P. gingivalis</i> by salivary flow. After further testing, it was discovered that the <i>S. cristatus </i> signaling activity was specific to the <i>fimA</i> gene, and is also associated with a 59-kDa surface protein. The size of the surface protein, however, suggests that it is inherently distinct from the signaling peptides that are secreted via other gram-positive bacteria [6]. | One such example of this involves the gram-negative, anaerobic bacteria <i>Porphyromonas gingivalis</i>. <i>P. gingivalis</i> is a predominant pathogen in severe cases of adult periodontitis and attaches to the tooth surface after the initial colonization of the gram-positive <i>Streptococcus</i> species. The colonization of <i>P. gingivalis</i> is key in the transformation of the bacterial biofilm from a commensal plaque to the pathogenic form observed in periodontitis [6]. Its colonization is also dependent on its attachment to oral surfaces, which is accomplished via a fimbriae-mediated adhesion. The <i>fimA</i> gene encodes for this subunit protein, which can be regulated environmentally [6]. Interestingly, the <i>P. gingivalis</i> <i>fimA</i> gene expression was found to be severely downregulated by the unrelated, gram-positive <i>Streptococcus cristatus</i> CC5A, which can be found in the initial colonization of dental biofilms. By interfering with the gene that encodes for the fimbriae-subunit protein, <i>S. cristatus</i> is able to prevent <i>P. gingivalis</i> fimbriae attachment to the biofilm, thus resulting in an easier removal of <i>P. gingivalis</i> by salivary flow. After further testing, it was discovered that the <i>S. cristatus </i> signaling activity was specific to the <i>fimA</i> gene, and is also associated with a 59-kDa surface protein. The size of the surface protein, however, suggests that it is inherently distinct from the signaling peptides that are secreted via other gram-positive bacteria [6]. | ||
Through these findings, it was clear that some signaling mechanisms between <i>P. gingivalis</i> and <i>S. cristatus</i> were occurring which was preventing the colonization of <i>P. gingivalis</i> on the plaque biofilm. Of course, it is likely that other microbial species also contribute to the upregulation of this gene, and thus may affect the downregulation of <i>fimA </i> caused by <i>S. cristatus</i>. It is also known that <i>Fusobacterium nucleatum</i> is involved with some type of cell-sigaling interaction with <i>P. gingivalis</i>, but in a different way. <i>F. nucleatum</i> is instead required for the <i>P. gingivalis</i> aggregation with the facultative aerobes already present on the biofilm [5]. This directly shows that inter-species communication between the different microbial species on dental plaque biofilm is required for proper biofilm maturation and is thus directly involved in oral disase propagation as well. This research should give us a better understanding of the regulation of <i>P. gingivalis</i> via cell-cell interactions within the dental plaque biofilm. | Through these findings, it was clear that some signaling mechanisms between <i>P. gingivalis</i> and <i>S. cristatus</i> were occurring which was preventing the colonization of <i>P. gingivalis</i> on the plaque biofilm. Of course, it is likely that other microbial species also contribute to the upregulation of this gene, and thus may affect the downregulation of <i>fimA </i> caused by <i>S. cristatus</i>. It is also known that <i>Fusobacterium nucleatum</i> is involved with some type of cell-sigaling interaction with <i>P. gingivalis</i>, but in a different way. <i>F. nucleatum</i> is instead required for the <i>P. gingivalis</i> aggregation with the facultative aerobes already present on the biofilm [5]. This directly shows that inter-species communication between the different microbial species on dental plaque biofilm is required for proper biofilm maturation and is thus directly involved in oral disase propagation as well. This research should give us a better understanding of the regulation of <i>P. gingivalis</i> via cell-cell interactions within the dental plaque biofilm. | ||
<br> | <br> | ||
Revision as of 23:58, 14 April 2009
By: Anna Frutiger
Introduction
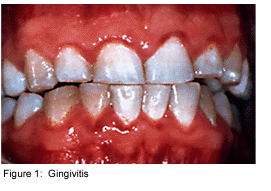
Dental plaque is commonly known as the primary cause of dental caries and other oral infections. Rightly so, as despite the increasing knowledge in oral health care, the average adult in the United States has between 10 and 17 decayed, missing or filled permanent teeth [4]. Additionally, the majority of the population has at least minor gingivitis, with a much smaller percentage suffering from moderate to severe periodontitis [4]. Dental plaque, which exists not only on the tooth surface but also under the gums, can be defined as a diverse community of microorganisms in the form of a biofilm. The microorganisms bind tightly to one another in addition to the solid tooth surface by means of an extracellular matrix consisting of polymers of both host and microbial origin [1][2]. Only recently has dental plaque begun to be considered a biofilm, which has further contributed to understanding periodontal disease, among others, and has helped to find the best ways to prevent and control this in the future [3].
As a biofilm, dental plaque has an open architecture much like that of other biofilms which consists of channels and voids, that help to achieve the flow of nutrients, waste products, metabolites, enzymes, and oxygen through the biofilm[4]. Because of this structure, a variety of microbial organisms can make up biofilms, including both aerobic and anaerobic bacteria. The microbial composition of dental biofilms includes over 700 species of bacteria and archaea, which all exist in a relatively stable environment called microbial homeostasis [2]. Dental plaque biofilms are responsible for many of the diseases common to the oral cavity including dental caries, periodontitis, gingivitis, and the less common periimplantitis (similar to periodontitis, but with dental implants), however biofilms are also present on healthy teeth as well [5].
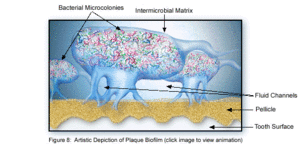
The microbial make-up of the normal plaque biofilm, however, differs immensely from the biofilm formed in carious regions and in the sub-gingival regions associated with periodontal disease. Because dental biofilms (and other biofilms) express an entirely different set of genes than free-floating bacteria, they are of particular interest to researchers and thus much is currently being done to explore the diverse nature of these microbial structures. The biofilm state brings about major changes in bacteria behavior such as interactions with each other and the host, and their response to environmental conditions [5]. Because of these changes, in addition to the microbial differences present among healthy and diseased biofilms, much of the research currently being done is in relation to plaque-related diseases and other health issues within the oral cavity. This is of major concern to the medical community, because it is being realized that oral pathologies have a major impact not only on the health of the oral cavity, but also on the overall health of the human body.
Formation of Dental Biofilms
The formation of dental plaque biofilms includes a series of steps that preferentially form at stagnant sites providing protection from the forces of removal. Plaque biofilms can exist on a variety of surfaces including fissues, smooth surfaces and gingival crevices, however they are most likely to be seen in their mature state in fissures and crevices, as these places are more difficult to reach with biofilm removal tools, like a toothbrush [1][5]. Additionally, through the growth process of the plaque biofilm, the microbial composition changes from one that is primarily gram-positive and streptococcus-rich to a structure filled with gram-negative anaerobes in its more mature state [6].
Adsorption of Host and Bacterial Molecules to the Tooth Surface
The first step in plaque biofilm development is the adsorption of host and bacterial molecules to the tooth surface. Within minutes of tooth eruption or a cleaning, pellicle formation begins, which is a thin coat of salivary proteins [3]. The pelicle acts like an adhesive by sticking to the tooth surface and encouraging a conditioning film of bacteria to attach to the pellicle[1]. This conditioning film directly influences the initial microbial colonization, and continues to adsorb bacteria to the tooth surface.
Healthy tooth surfaces and gingivae tend to only be associated with this first phase of biofilm development. It consists of an initial few layers (1-20) of mostly gram-positive cocci bacteria, followed by some gram-positive rods and fillaments and a very small amount of gram-negative cocci. The gram-positive cocci species involved in this conditioning layer include Streptococcus mutans, Streptococcus mitis, Streptococcus sanguis, Streptococcus oralis, Rothia dentocariosa, and Staphylococcus epidermidis. The gram-negative rod and filament species consist of Actinomyces viscosus, Actinomyces israelis. Actinomyces gerencseriae and the Corynebacterium species. Veillonella parvula and the Neisseria species make up the few gram-negative cocci, which are also aerobes or facultative aerobes and are able to adhere to the hard tooth surfaces which are non-exfoliating [5]. This early composition of the biofilm is able to withstand many of the frequent mechanisms of the oral cavity that contribute to bacterial removal such as swallowing, nose blowing, chewing, and salivary fluid outflow. The early colonizers are also able to survive in the high oxygen concentrations present in the oral cavity, without having much protection from other bacteria [5]. Thus, this thin, initial biofilm is almost always present on the tooth surface as it forms immediately after cleaning.
Passive Transport of Oral Bacteria to the Tooth Surface
Following pellicle formation, there is passive transport of oral bacteria to the tooth surface, which involves a reversible adhesion process [2]. By using weak, long-range physicochemical interactions between the pellicle coated tooth surface and the microbial cell surface, an area of weak attraction is formed that encourages the reversible adhesion [1]. This reversible adhesion then leads to a much stronger, irreversible attachment, as short-range interactions between specific molecules on the bacterial cells and the complementary receptor proteins on the pellicle surface occur. Because many oral bacteria have multiple adhesion types on their cell surface, they can thus participate in a plethora of interactions with both other bacteria and with the host surface molecules [1].
Co-Adhesion of later colonizers to already attached early colonizers
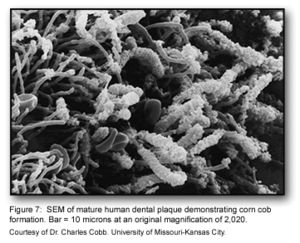
This stage of biofilm development continues to involve many specific interactions between bacterial receptors and adhesions. These interactions continue to build up the biofilm to create a more diverse environment. Further, the development of unusual morphological structures like corn-cobs and rosettes are formed during this phase [1].
The many interactions between these diverse bacterial species begin to create a number of synergistic and antagonistic biochemical interactions. For example, bacterial residing in food chains may help to contribute metabolically with other bacteria if they are physically located close to one another. Similarly, when obligate anaerobes and aerobes are involved in co-adhesion, these interactions can ensure the anaerobic bacteria’s survival in the oxygen-rich oral cavity [1].
Multiplication of the Attached Microorganisms
Eventually, the bacterial cells continue to divide until a three-dimensional mixed-culture biofilm forms that is spacially and functionally organized. Polymer production causes the development of the extracellular matrix, which consists of soluble and insoluble glucans, fructans, and heteropolymers [1]. This matrix is one of the key structural aspects of the plaque biofilm, much like that of other biofilms. Biofilms such as this are very thick, consisting of 100-300 cell layers. The bacterial stratification is arranged according to metabolism and aerotolerance, with the number of gram-negative cocci, rods and filaments increasing as more anaerobic bacteria appear [5]. As the biofilm thickens and becomes more mature, these anaerobic bacteria can live deeper within the biofilm, to further protect them from the oxygen rich environment within the oral cavity.
Inter-species Communication
The discovery that communication between cells in biofilm communities occurs has been key in understanding how dental plaque acts as a single unit. Communication can occur in a variety of ways, including gene expression, cell-cell signaling (ex. quorum sensing), and antibiotic resistance, among others. Specific bacteria within the biofilm community are able to act with other species to both help and impair the host, in addition to providing a positive cooperation between the different species of the biofilm [5]. Further, the patterns observed of microbial colonization and coaggregation appear to be primarily uni-directional, thus indicating that many of the bacterial species in dental biofilms require an environment that was previously habituated by other microbiota in order to properly colonize [5]. These specific cell-cell interactions have proven to be very important topics of current research involving dental plaque biofilms.
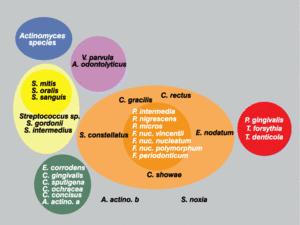
Sigmund Socransky and Anne Haffajee have been able to explain much of the unique colonization patterns and positive cooperation observed in plaque biofilm. The method that has helped explain this is one involving “clustered” groups of bacteria, where each cluster is created based on nutritional and atmospheric environments [5][7]. Each of the clusters is said to influence the other clusters, in addition to being related to a specific periodontal state, thus showing that these microbes are very closely associated with one another [5]. Socransky and Haffajee found that within different periodontal states, either all of none of the species belonging to a particular cluster were found. Very few individual species or pairs of species were found, which further helped show that biofilms were a community of microbes. Particular clusters were also found to interact particularly well with another cluster. For example, microbial species found in the red cluster were rarely observed without the presence of the orange cluster species [5]. Because of these associations found by Socransky and Haffajee and their cluster analysis, it provided further knowledge to idea that the microbial communities in plaque biofilms were involved in a great deal of inter-species communication.
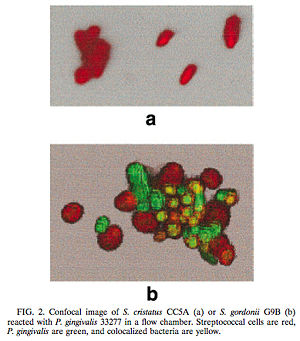
One such example of this involves the gram-negative, anaerobic bacteria Porphyromonas gingivalis. P. gingivalis is a predominant pathogen in severe cases of adult periodontitis and attaches to the tooth surface after the initial colonization of the gram-positive Streptococcus species. The colonization of P. gingivalis is key in the transformation of the bacterial biofilm from a commensal plaque to the pathogenic form observed in periodontitis [6]. Its colonization is also dependent on its attachment to oral surfaces, which is accomplished via a fimbriae-mediated adhesion. The fimA gene encodes for this subunit protein, which can be regulated environmentally [6]. Interestingly, the P. gingivalis fimA gene expression was found to be severely downregulated by the unrelated, gram-positive Streptococcus cristatus CC5A, which can be found in the initial colonization of dental biofilms. By interfering with the gene that encodes for the fimbriae-subunit protein, S. cristatus is able to prevent P. gingivalis fimbriae attachment to the biofilm, thus resulting in an easier removal of P. gingivalis by salivary flow. After further testing, it was discovered that the S. cristatus signaling activity was specific to the fimA gene, and is also associated with a 59-kDa surface protein. The size of the surface protein, however, suggests that it is inherently distinct from the signaling peptides that are secreted via other gram-positive bacteria [6].
Through these findings, it was clear that some signaling mechanisms between P. gingivalis and S. cristatus were occurring which was preventing the colonization of P. gingivalis on the plaque biofilm. Of course, it is likely that other microbial species also contribute to the upregulation of this gene, and thus may affect the downregulation of fimA caused by S. cristatus. It is also known that Fusobacterium nucleatum is involved with some type of cell-sigaling interaction with P. gingivalis, but in a different way. F. nucleatum is instead required for the P. gingivalis aggregation with the facultative aerobes already present on the biofilm [5]. This directly shows that inter-species communication between the different microbial species on dental plaque biofilm is required for proper biofilm maturation and is thus directly involved in oral disase propagation as well. This research should give us a better understanding of the regulation of P. gingivalis via cell-cell interactions within the dental plaque biofilm.
Section 3
Include some current research in each topic, with at least one figure showing data.
Conclusion
Overall paper length should be 3,000 words, with at least 3 figures.
References
1 Marsh, P.D. "Dental Plaque as a Microbial Biofilm." Caries Research. 2004. Volume 38. p.204-211.
2 Marsh, P.D. "Dental plaque as a biofilm and a microbial community - implications for health and disease." BMC Oral Health. 2006. 6(Suppl 1): S14.
3 Nield-Gehrig, J. S. "Dental Plaque Biofilms." Dental Plaque Biofilms.
4 Overman, P. R. "Biofilm: A New View of Plaque." The Journal of Contemporary Dental Practice. 2000. Volume 1. No. 3. P. 018.
5 Sbordone, L., Bortolaia, C. "Oral microbial biofilms and plaque-related diseases: microbial communities and their role in the shift from oral health to disease." Clin Oral Invest. 2003. Volume 7. P. 181-188.
6 Hua, X., Cook, G. S., Costerton, J. W., Bruce, G., Rose, T. M., Lamont, R. J. "Intergeneric Communication in Dental Plaque Biofilms." Journal of Bacteriology. 2000. Volume 182. p. 7067-7069.
7 Socransky, S. S., Haffajee, A. D., "Periodontal Microbial Ecology." Periodontology 2000. Volume 38. p. 135-187.
Edited by Anna Frutiger, student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.
