Desulfosarcina: Difference between revisions
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
'''NCBI:<br />[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=2299&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy]<br /> Genome ''' | '''NCBI:<br />[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=2299&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy]<br /> Genome ''' | ||
|} | |} | ||
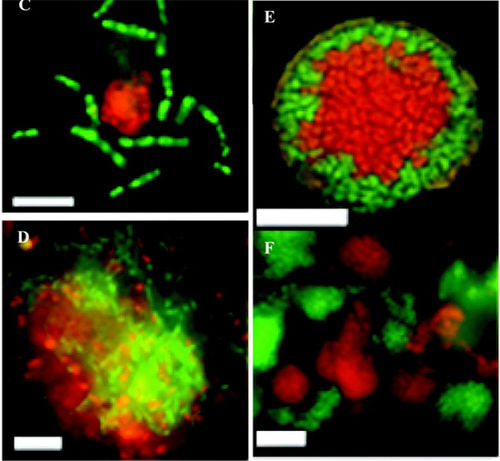
[[Image:desulfosarcina.jpg]] | [[Image:desulfosarcina.jpg|thumb|500px|right|''Desulfosarcina. ''"''(C'') Color overlay of archaeal ANME-1 rods visualized with the ANME1-862 probe labeled with fluorescein (in green), and ''Desulfosarcina ''spp. stained with the DSS_658 probe labeled with Cy-3 (in red). (''D'') Large archaeal ANME-1/sulfate-reducing ''Desulfosarcina ''aggregate, showing an apparently random association of the two groups. Scale bar = 10 µ :m. (''E'') Color overlay of a layered ANME-2/DSS aggregate showing a core of ANME-2 ''Archaea ''(hybridized with EelMSMX932 probe), surrounded by sulfate-reducing ''Desulfosarcina ''(hybridized with DSS658 probe) imaged by laser scanning confocal microscopy. (''F'') Large aggregation of loosely associated, individual microcolonies of ANME-2 (EelMSMX932_cy3), and ''Desulfosarcina ''(DSS658_FITC)." Image & Information courtesy of [http://www.arches.uga.edu/ Arches], specifically p.10 [http://www.arches.uga.edu/~whitman/coursedocs/mibo4300/natural populations and methods.PDF here.]]] | ||
==Classification== | ==Classification== | ||
===Higher order taxa:=== | ===Higher order taxa:=== | ||
Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae | |||
=== | ===Species:=== | ||
''Desulfosarcina variablis | ''Desulfosarcina variablis, Desulfosarcina sp. CME1'' | ||
==Description and Significance== | ==Description and Significance== | ||
''Desulfosarcina'' is a sulfate-reducing bacteria, and a member of the δ-proteobacteria. ''Desulfosarcina'' is a Group II sulfate-reducing bacteria because it can can utilize acetate and other fatty acids, oxidizing them completely. Other Group II sulfate-reducers include ''[http://biology.kenyon.edu/Microbial_Biorealm/bacteria/thermodesulfobacterium/thermodesulfobacterium.htm Desulfobacterium]'' and ''Desulfobacter''. | |||
Many sulfate-reducing bacteria are also members of the δ-proteobacteria, but have significant phylogenetic diversity. Despite this diversity, the analysis of their 16S rRNA has revealed that those in the δ-proteobacteria are more related to one another than to other bacteria. | Many sulfate-reducing bacteria are also members of the δ-proteobacteria, but have significant phylogenetic diversity. Despite this diversity, the analysis of their 16S rRNA has revealed that those in the δ-proteobacteria are more related to one another than to other bacteria. | ||
| Line 34: | Line 27: | ||
==Genome Structure== | ==Genome Structure== | ||
Though the Göttingen Genomics Laboratory has completed the genome sequences of ''Methanosarcina mazei'', ''Clostridium tetani'' and ''Thermus thermophilus'', and has stated that the genome of ''Desulfobacterium autotrophicum'' will also be finished soon, there is currently very little information concerning the genetics of ''Desulfosarcina''. | |||
==Cell Structure and Metabolism== | ==Cell Structure and Metabolism== | ||
[[Image:b.jpg]][[Image:d.jpg|frame|left|''D. variabilis'' contains significant quantities of AEG-P (top), and almost 20% of the alkyl-glycerol bond is present as DPG lipids, also known as "cardiolipin" (bottom). Note that the DPGs contain either ether or ester linkages in the core lipids.[http://www.nies.go.jp/biology/mcc/strainlist_a.htm#Anabaena][http://www.chemistry.montana.edu/chem524/pdf/Julian lipids RCM 2004.pdf Sturt ''et al'']]] | |||
[[Image:b.jpg]] | |||
</div> | </div> | ||
Revision as of 20:30, 2 June 2006
|
NCBI: |
Classification
Higher order taxa:
Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae
Species:
Desulfosarcina variablis, Desulfosarcina sp. CME1
Description and Significance
Desulfosarcina is a sulfate-reducing bacteria, and a member of the δ-proteobacteria. Desulfosarcina is a Group II sulfate-reducing bacteria because it can can utilize acetate and other fatty acids, oxidizing them completely. Other Group II sulfate-reducers include Desulfobacterium and Desulfobacter.
Many sulfate-reducing bacteria are also members of the δ-proteobacteria, but have significant phylogenetic diversity. Despite this diversity, the analysis of their 16S rRNA has revealed that those in the δ-proteobacteria are more related to one another than to other bacteria.
Sulfate-reducing bacteria fall into over 20 separate genera and these can be classified by the types of substrates that are utilized. A convenient division is by the ability to utilize acetate as a source of electrons. Group II is described above, and Group I sulfate reducers, such as Desulfovibrio, Desulfobacula and Desulfomicrobium, are not capable of utilizing acetate. These species only partially oxidize other organic substrates to acetate and excrete it into the medium.
A third group of sulfur-reducing organisms are unable to utilize sulfate, but can reduce sulfur to sulfide. These include genera such as Desulfuromonas and Desulfurella. They utilize substrates such as acetate or ethanol and couple this to the reduction of sulfur or other electrons acceptors such as thiosulfate, sulfite or dimethyl sulfoxide. This ability is not unique to these bacteria, but is widespread among chemoheteroorganotrophic bacteria.Salmonella, Campylobacter, and Proteus are all capable of this type of reduction; and for more information on this fascinating process please click here.
Genome Structure
Though the Göttingen Genomics Laboratory has completed the genome sequences of Methanosarcina mazei, Clostridium tetani and Thermus thermophilus, and has stated that the genome of Desulfobacterium autotrophicum will also be finished soon, there is currently very little information concerning the genetics of Desulfosarcina.
Cell Structure and Metabolism
| width="53%" | Sulfate reducers have a wide range of cellular morphologies, including rods, vibrios, ovals, spheres and even tear-dropped or onion shaped cells. Some are motile, others are not. Most sulfate-reducing bacteria are mesophilic, but a few thermophiles are known. Desulfosarcina variabilis is mesophilic, and contains bacterial core lipids (see images on left). The dominant phospholipid headgroups in D. variabilis are Phosphoethanolamine PE (48%) and Phosphoglycerol PG (33%). One study has found that Desulfosarcina variabilis solely contained n-hexadecyl ether side chains. For more information on tetraether lipids found in Archaea, click here. |}
Ecology
Although sulfate reduction is thought to be an anaerobic process, sulfate-reducing bacteria (SRB) are also important in aerobic environments if they can proliferate in anaerobic zones. For example, in marine sediments and in aerobic wastewater treatment systems, sulfate reduction accounts for up to 50% of the mineralization of organic matter. Furthermore, sulfate reduction strongly stimulates microbially enhanced corrosion of metals. {| width="100%" |

The Plum Island Estuary Microbial Observatory (PIMO), located at the Plum Island Estuary LTER site in coastal Massachusetts, identifies prokaryotes in salt marsh sediments and plankton and determines their role in controlling major ecosystem processes. Among SRBs identified, relatives of Desulfosarcina variabilis and Desulfobacterium anilini were found to be persistent in the sediment.
|- | height="21" |
|}
