Domestic cat intestinal microbiome: Difference between revisions
| Line 12: | Line 12: | ||
In the gastrointestinal tract of normal, healthy domestic cats, there are five major bacterial phyla present. These are predominantly classified as <i>Firmicutes</i> (68%), followed by<i> Proteobacteria</i> (14%), <i>Bacteroidetes</i> (10%), <i>Fusobacteria</i> (5%), and <i>Actinobacteria</i> (4%). Within the <i>Firmicutes</i> phylum, the majority of bacteria belong to the orders <i>Clostridiales</i> (54%), followed by <i>Lactobacillales</i> (14%),<i> Bacteroidales</i> (11%), <i>Campylobacterales</i> (10%), and <i>Fusoacteriales</i> (6%). Specifically, within the <i>Clostridiales</i> order, the predominant domains are <i>Clostridium clusters I</i> (58%) and <i>XIVa</i> (27%).The intestinal microbiota of the healthy specific pathogen-free cat showed a decreased bacterial diversity when compared to normal raised domestic cat, with 98% of all bacterial classified within the phylum <i>Firmicutes</i>.<ref name=Cat_type>[https://pubmed.ncbi.nlm.nih.gov/11149714/=Ritchie, Lauren E et al. “Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis.” <i>FEMS microbiology ecology</i> vol. 66,3 (2008): 590-8. doi:10.1111/j.1574-6941.2008.00609.x]</ref> | In the gastrointestinal tract of normal, healthy domestic cats, there are five major bacterial phyla present. These are predominantly classified as <i>Firmicutes</i> (68%), followed by<i> Proteobacteria</i> (14%), <i>Bacteroidetes</i> (10%), <i>Fusobacteria</i> (5%), and <i>Actinobacteria</i> (4%). Within the <i>Firmicutes</i> phylum, the majority of bacteria belong to the orders <i>Clostridiales</i> (54%), followed by <i>Lactobacillales</i> (14%),<i> Bacteroidales</i> (11%), <i>Campylobacterales</i> (10%), and <i>Fusoacteriales</i> (6%). Specifically, within the <i>Clostridiales</i> order, the predominant domains are <i>Clostridium clusters I</i> (58%) and <i>XIVa</i> (27%).The intestinal microbiota of the healthy specific pathogen-free cat showed a decreased bacterial diversity when compared to normal raised domestic cat, with 98% of all bacterial classified within the phylum <i>Firmicutes</i>.<ref name=Cat_type>[https://pubmed.ncbi.nlm.nih.gov/11149714/=Ritchie, Lauren E et al. “Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis.” <i>FEMS microbiology ecology</i> vol. 66,3 (2008): 590-8. doi:10.1111/j.1574-6941.2008.00609.x]</ref> | ||
[[Image:Catduodenum.png |thumb|300px|left|<b>Figure 3:</b> Electron micrograph of a portion of the duodenum of a healthy cat. [https://pubmed.ncbi.nlm.nih.gov/11149714/].]] | [[Image:Catduodenum.png |thumb|300px|left|<b>Figure 3:</b> Electron micrograph of a portion of the duodenum of a healthy cat. [https://pubmed.ncbi.nlm.nih.gov/11149714/].]] | ||
Approximately four-fifths of all bacteria found were classified as <i>Firmicutes</i>, exhibiting diverse representation across various <i>Clostridium</i> clusters within the <i>Clostridiales</i> class. These bacteria were affiliated with six different <i>Clostridium</i> clusters: clusters I, III, IV, XI, XIVa, and XIVb. Roughly half of these <i>Firmicutes</i> were found in the jejunum, with slightly fewer in the colon, and the rest in the ileum.<ref name=Cat_type> | |||
<i>Actinobacteria</i> distribution was fairly evenly split, with approximately one-third found in the ileum, another third in the colon, and the remaining third in the jejunum.<ref name=Cat_type> | |||
Regarding <i>Bacteroidetes</i>, about half of them were located in the ileum, with a significant portion in the colon and a smaller proportion in the rectum. The jejunum had only a negligible amount of <i>Bacteroidetes</i>.<ref name=Cat_type> | |||
The majority of <i>Fusobacteria</i>, roughly two-thirds, were found in the colon, followed by smaller proportions in the jejunum and ileum.<ref name=Cat_type> | |||
The phylum <i>Proteobacteria</i> showed a varied distribution. Approximately one-third of <i>Proteobacteria</i> clones were found in the duodenum, ileum, and colon each, with smaller proportions in the jejunum and rectum.<ref name=Cat_type> | |||
In the duodenum of healthy domestic cats, the levels of total bacteria are consistent, with a dominant presence of anaerobic bacteria like <i>Clostridium</i> and <i>Bacteroides</i>, which are known for their ability to deconjugate bile acids and bind cobalamin, essential for digestive health. Domestic cats with clinical symptoms of gastrointestinal issues had lower counts of several bacteria species, including <i>Pasteurella</i>, <i>Bacteroides</i>, and <i>Lactobacillus</i>. <i>Pasteurella</i> are commonly found in the respiratory tract but can also inhabit the gastrointestinal tract. <i>Bacteroides</i>, which plays a crucial role in bile acid metabolism and is associated with steatorrhea, a condition characterized by fat malabsorption. <i>Lactobacillus</i> are beneficial bacteria known for their role in maintaining gut health and aiding in digestion.<ref name=Cat_duodenum>[https://pubmed.ncbi.nlm.nih.gov/11149714/=Johnston, K L et al. “Comparison of the bacterial flora of the duodenum in healthy cats and cats with signs of gastrointestinal tract disease.” <i>Journal of the American Veterinary Medical Association</i> vol. 218,1 (2001): 48-51. doi:10.2460/javma.2001.218.48]</ref> | In the duodenum of healthy domestic cats, the levels of total bacteria are consistent, with a dominant presence of anaerobic bacteria like <i>Clostridium</i> and <i>Bacteroides</i>, which are known for their ability to deconjugate bile acids and bind cobalamin, essential for digestive health. Domestic cats with clinical symptoms of gastrointestinal issues had lower counts of several bacteria species, including <i>Pasteurella</i>, <i>Bacteroides</i>, and <i>Lactobacillus</i>. <i>Pasteurella</i> are commonly found in the respiratory tract but can also inhabit the gastrointestinal tract. <i>Bacteroides</i>, which plays a crucial role in bile acid metabolism and is associated with steatorrhea, a condition characterized by fat malabsorption. <i>Lactobacillus</i> are beneficial bacteria known for their role in maintaining gut health and aiding in digestion.<ref name=Cat_duodenum>[https://pubmed.ncbi.nlm.nih.gov/11149714/=Johnston, K L et al. “Comparison of the bacterial flora of the duodenum in healthy cats and cats with signs of gastrointestinal tract disease.” <i>Journal of the American Veterinary Medical Association</i> vol. 218,1 (2001): 48-51. doi:10.2460/javma.2001.218.48]</ref> | ||
Revision as of 00:28, 14 April 2024
Introduction
By Xinyi Liu

Felines have been domesticated for a long time, becoming cherished companions for humans. Similar to humans, they are colonized by bacteria during birth. When kittens are born, they are exposed to the external environment, leading to changes in their gut microbiota. After weaning, felines consume high-protein foods in large quantities, causing significant changes in the species composition and structure of their gut microbiota.The differing diets of humans and felines contribute to significant variations in the composition of their intestinal microbiota.[1] These gut microbes play crucial roles in the immune and digestive systems of felines. Given their prolonged coexistence with humans, the condition of domestic pet cats' gut microbiota can impact not only their own health but also that of their owners.[2]
Composition of domestic cat intestinal microbiome
The feline gastrointestinal tract harbors a complex ecosystem comprising various microorganisms, including bacteria, viruses, fungi, and protozoa. Among these, bacteria constitute over 98% of the population, and predominantly strict and facultative anaerobic bacteria dominate the gut microbiota. [3]Their metabolic activities generate beneficial molecules like food or drug compounds, enhancing the host's immune response and metabolic capabilities. These intestinal bacteria play a crucial role in regulating the immune system, defending against external pathogens, and supplying essential nutrients, such as vitamins that may be scarce or challenging for the host to produce independently. Gut bacteria in cats play a crucial role not only in digestive diseases like inflammatory bowel disease or alimentary small cell lymphoma but also in the overall well-being of an organism.[4] While the precise mechanisms are still under investigation, it's clear that gut bacteria can impact a wide range of diseases affecting various organs and systems, including but not limited to diabetes, chronic kidney disease, and obesity.[5][6][7]
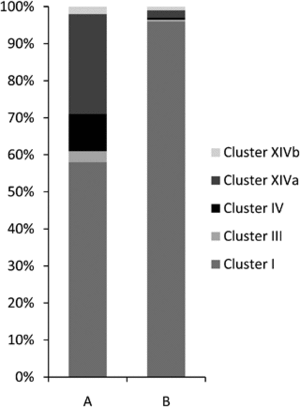
In the gastrointestinal tract of normal, healthy domestic cats, there are five major bacterial phyla present. These are predominantly classified as Firmicutes (68%), followed by Proteobacteria (14%), Bacteroidetes (10%), Fusobacteria (5%), and Actinobacteria (4%). Within the Firmicutes phylum, the majority of bacteria belong to the orders Clostridiales (54%), followed by Lactobacillales (14%), Bacteroidales (11%), Campylobacterales (10%), and Fusoacteriales (6%). Specifically, within the Clostridiales order, the predominant domains are Clostridium clusters I (58%) and XIVa (27%).The intestinal microbiota of the healthy specific pathogen-free cat showed a decreased bacterial diversity when compared to normal raised domestic cat, with 98% of all bacterial classified within the phylum Firmicutes.[8]
Approximately four-fifths of all bacteria found were classified as Firmicutes, exhibiting diverse representation across various Clostridium clusters within the Clostridiales class. These bacteria were affiliated with six different Clostridium clusters: clusters I, III, IV, XI, XIVa, and XIVb. Roughly half of these Firmicutes were found in the jejunum, with slightly fewer in the colon, and the rest in the ileum.Cite error: Closing </ref> missing for <ref> tag
Section 2
Include some current research, with at least one figure showing data.
Section 3
Include some current research, with at least one figure showing data.
Section 4
Section 5
Section 6
Conclusion
References
- ↑ Lubbs, Dustin C. et al. "Dietary protein concentration affects intestinal microbiota of adult cats: a study using DGGE and qPCR to evaluate differences in microbial populations in the feline gastrointestinal tract.” Journal of animal physiology and animal nutrition 93 1 (2009): 113-21.
- ↑ Alessandri, Giulia et al. “Catching a glimpse of the bacterial gut community of companion animals: a canine and feline perspective.” Microbial biotechnology vol. 13,6 (2020): 1708-1732. doi:10.1111/1751-7915.13656
- ↑ Lauren E et al. “Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis.” FEMS microbiology ecology vol. 66,3 (2008): 590-8. doi:10.1111/j.1574-6941.2008.00609.x
- ↑ Ganz, Holly H et al. “The Kitty Microbiome Project: Defining the Healthy Fecal "Core Microbiome" in Pet Domestic Cats.” Veterinary sciences vol. 9,11 635. 16 Nov. 2022, doi:10.3390/vetsci9110635
- ↑ Kieler, Ida Nordang, et al. "Diabetic cats have decreased gut microbial diversity and a lack of butyrate producing bacteria." Scientific reports 9.1 (2019): 4822.
- ↑ Summers, Stacie C et al. “The fecal microbiome and serum concentrations of indoxyl sulfate and p-cresol sulfate in cats with chronic kidney disease.” Journal of veterinary internal medicine vol. 33,2 (2019): 662-669. doi:10.1111/jvim.15389
- ↑ Kieler, I N et al. “Overweight and the feline gut microbiome - a pilot study.” Journal of animal physiology and animal nutrition vol. 100,3 (2016): 478-84. doi:10.1111/jpn.12409
- ↑ Lauren E et al. “Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis.” FEMS microbiology ecology vol. 66,3 (2008): 590-8. doi:10.1111/j.1574-6941.2008.00609.x
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski,at Kenyon College,2024
