Drug Resistance in Mycobacterium Tuberculosis
Tuberculosis (TB) is a potentially deadly disease caused by pathogenic bacteria, usually Mycobacterium tuberculosis. It has existed in humans since ancient times and had high mortality rates without adequate treatment options before the invention of antibiotics , specifically streptomycin in 1943, that were potent enough to kill the bacteria. [6] In the 1960’s, following a drastic reduction in TB rates around the world, people began to predict that the disease could be completely eradicated within a century. [5] However, this goal proved overly-optimistic as drug-resistant strains had begun to emerge since the first use of antibiotics to treat TB. At first this was mainly due to only using a single drug, streptomycin, to treat the infection, prompting the use of multi-drug therapy but in recent decades multi-drug resistant (MDR), extensively- drug resistant (XDR), and totally-drug resistant (TDR) strains of TB have emerged. [1] Many of these strains are effectively incurable, especially the XDR and TDR strains, even for patients with access to an array of anti-TB drugs. [1] Given their grave public health threat it is crucial to study the molecular biology of the intrinsic and acquired mechanisms of resistance in M. tuberculosis in order to develop new drugs that avoid these mechanisms.
Intrinsic Drug Resistance
M. tuberculosis possess a multitude of resistance mechanisms against a wide range of antibiotics, as far as its intrinsic mechanisms (as opposed to acquired mechanisms that are brought about by chromosomal mutations, as discussed below) they can be divided into two categories: passive and specialized resistance. [7]
Passive Resistance Mechanisms
The primary feature of passive resistance to antibiotics in M. tuberculosis is a highly impermeable cell wall. Hydrophobic chemicals are unable to enter this cell wall due to a layer of hydrophilic arabinogalactan [6]. This layer is then wrapped in hydrophobic mycolic acids which severely limits the entrance of hydrophilic molecules. The mycolic acids are added to arabinogalactan in the cell wall by group of mycolyltransferase enzymes. One gene that encodes one of the mycolytransferases is the FbpA gene and studies of FbpA mutants have led to the conclusion that there is a strong connection between mycolic acid content and antibiotic resistance. [6] Despite being considered Gram-positive, Mycobacterium cell wall layers create space that resembles the periplasm of Gram-negative bacteria. [6] Another piece of evidence supporting the impermeability of the Mycobacterial cell wall is the fact that the time it takes for β-lactam antibiotics to diffuse through the cell wall takes a hundred times longer than it does for Escherichia coli. [7]
Specialized Resistance Mechanisms
The cell wall of mycobacteria is only part of what makes this species innately resistant to such a broad range of antibiotics. Studies have shown that the mycobacterial cell envelope lets in enough hydrophilic antibiotics over time to potentially kill the cell; however, since the cell wall significantly hinders the entrance of antibiotics it is thought that it slows antibiotic accumulation enough to allow for various cellular systems to detoxify the invading drugs. [6] This is especially evident in the case of β-lactams, which work by inhibiting the assembly of peptidoglycan and ultimately causing cell death, as peptidoglycan is a crucial part of the bacterial cell wall. [7] While the mycobacterial cell wall greatly limits the rate at which β-lactams are able to accumulate in the cell, effectively protecting against certain antibiotics, such as carbapenems which are not very stable and degrade relatively quickly, they still would be able to high enough levels over time due to the slow rate at which M. tuberculosis undergoes cell division. [7] However, M. tuberculosis possesses certain enzymes called β-lactamases that effectively degrade β-lactam antibiotics. Clinical evidence and in vitro experiments involving successful treatments of both β-lactams and β-lactamase inhibitors, as well as β-lactamase-resistant- β-lactams have strengthened the claim that β-lactamases are the most important factor in mycobacterial resistance to β-lactams. [6]
Another mechanism of specialized intrinsic resistance in M. tuberculosis is the molecular mimicry of certain drug targets by proteins. For example in the case of fluoroquinolones (fluoridated quinolones), which lead to cell death by preventing transcription, replication, and repair of DNA by interfering with DNA gyrase, a certain protein, MfpA, has been found to cause intrinsic resistance to fluoroquinolones. [6] It is able to bind to DNA gyrase, inhibiting its normal activity, and this is considered DNA mimicry as MfpA is similar in shape, size, and electrostatic nature to the double-helix of DNA. [11]
A third key mechanism of intrinsic drug resistance in M. tuberculosis is the presence of a multitude of drug efflux pumps that are able to expel antibiotics from the cell. In general efflux pumps are membrane-spanning proteins that are involved in transporting waste, nutrients, and other molecules across the cell wall. While deportation of certain antibiotics may be due to non-specific transportation and thus not an inherent purpose of efflux pumps, the presence of regulatory systems for these pumps that are believed to have evolved to increase antibiotic resistance makes drug efflux pumps a mechanism of specialized drug resistance. [7] In a study by Dinesh et al. [4] they looked at intrinsic resistance of M. tuberculosis efflux pumps to peptidoglycan synthesis inhibitors (PSI) with their results confirming that efflux pumps played a role in resistance to certain PSI including β-lactams, vancomycin, and bacitracin.
As of 2012 there were 18 different transporters that were characterized to be involved in antibiotic resistance or susceptibility of mycobacteria. [12] Two examples of these are IniBAC, causing resistance to isoniazid and ethambutol, and EfpA, which is a non-specific multi-drug transporter and returning to the concept of regulatory systems that have evolved to be responsive to the presence of antibiotics, the expression of both of these proteins is negatively controlled by a transcription regulator Lsr2 which is inducible by ethambutol or isoniazid. [7] A very recent study by Li et al. [13] examined gene expression of efflux pumps in clinically isolated strains of M. tuberculosis.
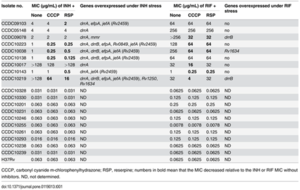
Table 1. Minimum inhibitory concentrations (MICs) for isoniazid (INH) and rifampicin (RIF) in the absence or presence of efflux inhibitors and the number of genes overexpressed in nine multidrug-resistant (MDR) M. tuberculosis isolates. CCCP (0.5 μg/mL) and reserpine (5 μg/mL) are both efflux pump inhibitors Li et al., 2015[13
] Their results (
Table 1. ) of both differences in minimum inhibitory concentrations (MICs) for isoniazid (INH) and rifampicin (RIF) when the cells were exposed or not exposed to efflux inhibitors showed no difference in MICs for pan-sensitive strains (susceptible to all standard antibiotics) while drug resistant strains exhibited considerable differences based on exposure to efflux inhibitors. Descriptions of which genes were overexpressed in MDR strains, led to the conclusion that efflux pumps likely play an important role in acquired INH resistance. [13]
Acquired Resistance
While in many other types of bacteria drug resistance is gained through horizontal gene transfer by plasmids or transposons, in M. tuberculosis all strains with acquired resistance that are currently known are through chromosomal mutations due to the selective pressure of antibiotics. [1] This selection of drug-resistant strains could not occur without the extensive and prolonged use of antibiotics necessary to treat the disease as those strains would have lower fitness under normal conditions. [7] Additionally recent studies, such as that by Kohanski et al. [10], have shown that sub-lethal doses of antibiotics can cause multi-drug resistance in E. Coli due to increased mutation rates by free radicals. This could very well apply to M. tuberculosis as well, suggesting that current TB drugs are not only selecting for drug-resistant strains but quite possibly creating them as well. There is a vast array of known mutations in the M. tuberculosis genome that have been shown to confer resistance to certain antibiotics. Knowledge of these mutations is crucial for improving the rapid detection of drug resistant strains. One example can be found in a study by Cui et al [9] in which they analyzed acquired resistance to Ethambutol, a first-line drug that works by inhibiting the synthesis of arabinogalactan in the cell wall, which has been gradually increasing in certain regions, by focusing on the intergenic region (IGR) between the embC and embA genes. They analyzed over seven hundred different strains for drug susceptibility and sequenced the embC-embA IGR and analyzed its transcriptional activity and ability to bind to EmbR, a transcription regulator of embAB. Previous studies have found that overexpression of the embAB gene is associated with high levels of Ethambutol-resistant [http://en.wikipedia.org/wiki/Arabinosyltransferase arabinosyltransferase activity. The data from this study shows that there was indeed a significant correlation between embC-embA IGR mutations and Ethambutol resistance and when using these mutations as markers for molecular drug susceptibility testing they found it significantly increased the sensitivity of the tests. [9]
Promising New Drugs and Drug Targets
Dinitrobenzamide derivatives (DNB)
In a study by Christophe et al. [2] they used confocal fluorescence microscopy to conduct an assay on the phenotypic effects of various chemical compounds on M. tuberculosis. After infecting macrophages with mycobacteria that expressed Green fluorescent protein (GFP) they monitored both amount of green fluorescent bacteria and host cell number. They used known anti-tuberculosis drug such as isoniazid and rifampin as controls and used high-throughput screening to test over 50,000 synthetic chemicals. Ultimately 135 were identified as potent inhibitors of mycobacteria in cells with no toxic effect on the host cells.
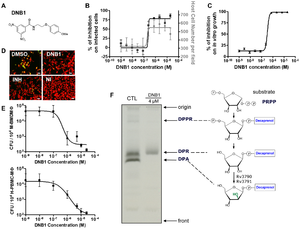
Figure 1. Activity Confirmation and Target Identification for N-(2-(4-methoxyphenoxy) ethyl)-3,5-dinitrobenzamide (DNB1) (A) Chemical structure of DNB1 (B) Results of intracellular assay from DNB1 of the benzamide scaffold (C) Inhibition of the in vitro growth fluorescence assays by DNB1 (D) Images of human macrophages infected with M. tuberculosis containing GFP after exposure to DNB1 and INH (5 µM) and DMSO for 7 days. NI: non infected (E) Dose-response curve for DNB1 on mouse macrophages infected with M. tuberculosis containing GFP and human macrophages after 5 days for 2 independent experiments (F) Gel showing the effect of DNB1 on the synthesis of decaprenyl-phospho-arabinose (DPA) by cell-free extracts of Mycobacterium smegmatis. Source: Christophe et al., 2009 [2
Of these compounds dinitrobenzamide derivatives (DNB) were highly effective against M. tuberculosis (
Figure 1. ) even against XDR strains.
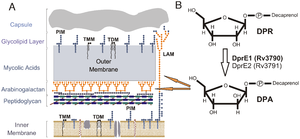
Figure 2. Representation of the Mycobacterial cell wall and pathway for DPA biosynthesis (A) Abbreviations: phosphatidylinositol mannosides (PIM), lipoarabinomannans (LAM), trehalose monomycolates (TMM), trehalose dimycolates (TDM) Arabinose sugars shown in orange (B) Formation of decaprenylphosphoryl arabinose (DPA) by the epimerization of decaprenylphosphoryl ribose (DPR) by DprE1 (Rv3790) and DprE2 (Rv3791). Orange arrows show contribution of DPA to arabinogalactan and LAM assembly Source: Crellin et al., 2011 [3
Their data showed that this was due to inhibition of the synthesis of the arabinogalactan and lipoarabinomannan, as DNB inhibits DprE1, which is part of decaprenylphosphoryl arabinose (DPA) necessary for arabinose donation for mycobacterial cell wall biosynthesis [2] [3] Nitro-benzothiazinones (BTZ) have also been identified as a potent new class of anti-mycobacterial drugs and like DNB they have been shown to target DprE1 and thus prevent the formation of DPA (
Figure 2) [3] . Perhaps the most important aspect of this recently discovered class of drugs is that they have the potential to inhibit M. tuberculosis replication in macrophages (
Figure 1D-F), as M. tuberculosis replication and survival in macrophages is an essential aspect of its success. [2]
Further Reading
As more and more highly drug-resistant strains of M. tuberculosis appear around the world it is crucial that more resources are invested in research on the molecular mechanisms of resistance in M. tuberculosis in order to identify new drug targets, novel antibiotics, and better means of rapid detection. It is even more pressing that we develop new tools to fight M. tuberculosis in the context of TDR strains that are virtually untreatable with today’s drugs. Yet it is also essential that the biochemical side of TB treatment is not separated from focus on the social aspects of fighting the disease, as huge numbers of people in impoverished and otherwise marginalized communities have little to no access to any sort of diagnosis or treatment for TB. Below are some resources for those interested in learning more about the ongoing battle against TB. Chapter on drug-resistant TB in Russian Prisons in Pathologies of Power by Paul Farmer (2003) A database that contains information on known genetic mutations in M. tuberculosis that confer drug resistance in which you can search by drug class: https://tbdreamdb.ki.se/Data/DrugArea.aspx?AreaID=AMI
References
3. Crellin PK, Brammananth R, Coppel RL (2011) Decaprenylphosphoryl-β-D-Ribose 2′-Epimerase, the Target of Benzothiazinones and Dinitrobenzamides, Is an Essential Enzyme in Mycobacterium smegmatis. PLoS ONE 6(2): e16869. doi:10.1371/journal.pone.0016869 4. Dinesh N, Sharma S, Balganesh M. Involvement of Efflux Pumps in the Resistance to Peptidoglycan Synthesis Inhibitors in Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy. 2013;57(4):1941-1943. doi:10.1128/AAC.01957-12.
9. Cui Z, Li Y, Cheng S, Yang H, Lu J, Hu Z, Ge B. Mutations in the embC-embA intergenic region contribute to mycobacterium tuberculosis resistance to ethambutol. Antimicrob Agents Chemother 2014 NOV;58(11):6837-43. 10. Kohanski MA, DePristo MA, Collins JJ. Sub-lethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Molecular cell. 2010;37(3):311-320. doi:10.1016/j.molcel.2010.01.003. 11. Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, Blanchard JS. 2005. A fluoroquinolone resistance protein from mycobacterium tuberculosis that mimics DNA. Science 308(5727):1480-3. 12. Viveiros M, Martins M, Rodrigues L, Machado D, Couto I, Ainsa J, Amaral L Inhibitors of mycobacterial efflux pumps as potential boosters for anti-tubercular drugs. Expert Rev Anti Infect Ther. 2012 Sep; 10(9):983-98. 13. Li G, Zhang J, Guo Q, Jiang Y, Wei J, et al. (2015) Efflux Pump Gene Expression in Multidrug-Resistant Mycobacterium tuberculosis Clinical Isolates. PLoS ONE 10(2): e0119013. doi:10.1371/journal.pone.0119013 Edited by (Noah Knowlton-Latkin), a student of Nora Sullivan in BIOL168L (Microbiology) in The Keck Science Department of the Claremont Colleges Spring 2014.
