Dyadobacter fermentans: Difference between revisions
| (One intermediate revision by the same user not shown) | |||
| Line 6: | Line 6: | ||
NCBI: | {| | ||
| height="10" bgcolor="#FFDF95" | | |||
'''NCBI: [https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Undef&id=120831&lvl=3&lin=s&keep=1&srchmode=1&unlock&log_op=lineage_toggle Taxonomy]''' | |||
|} | |||
''Dyadobacter fermentans'' | ''Dyadobacter fermentans'' | ||
The ''Dyadobacter'' Genus currently consists of 14 recognized species. They have been isolated from various environments such as freshwater, seawater, glaciers, desert sand, plants, soils, and animals. Its 16s rRNA gene sequence has less than 88% similarity to any other bacterial genus. | The ''Dyadobacter'' Genus currently consists of 14 recognized species. They have been isolated from various environments such as freshwater, seawater, glaciers, desert sand, plants, soils, and animals. Its 16s rRNA gene sequence has less than 88% similarity to any other bacterial genus. | ||
Latest revision as of 03:30, 26 April 2024
Classification
Higher order taxa
Bacteria; Bacteroidota; Cytophagia; Cytophagales; Spirosomaceae
Species
|
NCBI: Taxonomy |
Dyadobacter fermentans
The Dyadobacter Genus currently consists of 14 recognized species. They have been isolated from various environments such as freshwater, seawater, glaciers, desert sand, plants, soils, and animals. Its 16s rRNA gene sequence has less than 88% similarity to any other bacterial genus.
D. fermentans was the first species to be discovered in the Dyadobacter genus. It was first isolated by Marisa K. Chelius and Eric W. Triple in 2000, where the bacteria could not be assigned to any known genus, so it was consequently categorized as a new genus and species. D. fermentans is branched equidistant from the aquatic bacteria Runella slithyformis and Microscilla furvenscens. https://microbewiki.kenyon.edu/index.php?title=Dyadobacter_fermentans&action=info Species NCBI: Taxonomy
Genus species
Description and Significance
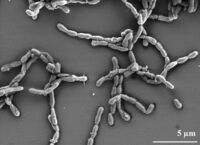
Dyadobacter fermentans is a Biosafety level 1 organism. It is aerobic, heterotrophic, and non-motile. It produces a pigment similar to flexirubin. It was discovered by Marisa K. Chelius and Eric W. Triplet in 2000. Isolated and discovered in 2000, Dyadobacter fermentans is an aerobic, gram-negative, non-motile, rod shaped chemoorganotroph. The rods can appear as pairs or in chains. D. fermentans is found in many plants, the cysts of nematodes (Heterodera glycines), and in contaminated soil. The term “fermentans” is derived from the bacterium's ability to ferment glucose and fructose. It can survive in nitrogen limited conditions by competing with the host plant for resources. This strain can also be isolated from the stem of maize plants when watered with fertilizer containing no nitrogen. When D. fermentans is found in Z. mays, the plants are healthy despite being nitrogen deficient. D. fermentans cannot survive in Z. mays when normal levels of nitrogen are present in the surrounding soil. This symbiosis between Z. mays and D. fermentans could have potential applications in agricultural practices.
Genome Structure
Dyadobacter fermentans has a genome that is 6,967,790 bp long that consists of one main circular chromosome. The chromosome has a 51.4% GC content. It has 5,854 predicted genes. Of the 5,854 genes, 5,804 genes were protein-coding and 50 were RNA-coding. [3] 64.7% of the protein-coding genes are thought to have a putative function, while the remaining protein-coding genes are designated as hypotehical. 85 pseudogenes have also been identified in the D. fermentans genome.
Cell Structure, Metabolism and Life Cycle
This bacterium’s carbon source is from carbohydrates, but not polymers. Its energy source is glucose and sucrose via fermentation. It has two membranes. D. fermentans is composed of unsaturated and saturated iso-branched species, straight chain unsaturated species, as well as iso-branched, hydroxylated species. The cells of D. fermentans are composed of polyamine and putrescine as major components and cadaverine and spermine as minor compents. Its main fatty acids include 43±5% 15: 0iso 2-OH}16: 1ω7c, 17±5% 16: 1ω5c and 16±8% 15: 0iso.
D. fermentans does not hydrolyze cellulose, agar, gelatin, or starch. However, it is able to produce acid from ribose. D. Fermentans is able to grow on acetate, D-arabitol, L-arabitol, amidon, fumarate, 5-ketogluconate, glucose, glycerol, D-lyxose, malate, malonate, sucrose, tartate, erythritol, D-glucose, D-fructose, mannose, L-sorbose, rhamnose, dulcitol, inositol, mannitol, sorbitol, methyl α-D-mannoside, methyl α-D-glucoside, N-acetylglucosamine, amygdaline, arbutine, aesculin, salicin, cellobiose, maltose, lactose, melibiose, saccharose, trehalose, inulin, melezitose, D-raffinose, xylitol, β-gentiobiose, D-turanose, D-tagatose, D-fucose, and L- fucose.
Ecology and Pathogenesis
Zea mays is a host to Dyadobacter fermentans, where it dwells in the roots of the plant. The presence or lack of D. fermentans has no significant influence over yield in Maize plants. Consequently, D. fermentans is thought to be neither beneficial nor pathogenic to maize. D. fermentans can decrease N2O emissions in soil, allowing it to be utilized as a bacterial form of denitrification in the rhizosphere. This ability has potential for various environmental applications, such as mitigating greenhouse gasses from agricultural fields. The lack of effect on dry yield means that D. fermentans could help maize grow in nitrogen deficient conditions.
Additionally, D. fermentans has been found in soybean nematodes (Heterodera glycines). However, the mechanisms of this potential symbiosis are largely unkown.
D. fermentans is not pathogenic.
References
[1] BacDive. (n.d.). Dyadobacter fermentans NS 114, NS114: Type strain: DSM 18053, ATCC 700827, CIP 107007, KCTC 52180: BacDiveID:3776. https://bacdive.dsmz.de/strain/3776
[2] Chelius, M. K., & Triplett, E. W. (2000, March 1). Dyadobacter fermentans gen. Nov., sp. nov., a novel Gram-negative bacterium isolated from surface-sterilized Zea mays stems. microbiologyresearch.org. https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/00207713-50-2-751
[3] Lang, E., Lapidus, A., Chertkov, O., Brettin, T., Detter, J. C., Han, C., Copeland, A., Glavina Del Rio, T., Nolan, M., Chen, F., Lucas, S., Tice, H., Cheng, J.-F., Land, M., Hauser, L., Chang, Y.-J., Jeffries, C. D., Kopitz, M., Bruce, D., … Klenk, H.-P. (2009, September 24). Complete genome sequence of Dyadobacter Fermentans type strain (NS114). Standards in genomic sciences. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3035232/
[4] Usyskin-Tonne, A., Hadar, Y., & Minz, D. (2019, May 20). Altering N2O emissions by manipulating wheat root bacterial community. Nature News. https://www.nature.com/articles/s41598-019-44124-3
[5] Shen L, Liu Y, Wang N, Adhikari NP. Genomic Insights of Dyadobacter tibetensis Y620-1 Isolated from Ice Core Reveal Genomic Features for Succession in Glacier Environment. Microorganisms. 2019; 7(7):211. https://doi.org/10.3390/microorganisms7070211
[6] Gao, J., Sun, P., Wang, X., Qiu, T., Lv, F., Yuan, M., Yang, M., & Sun, J. (2016). Dyadobacter endophyticus sp. nov., an endophytic bacterium isolated from maize root. International Journal of Systematic and Evolutionary Microbiology, 66(10), 4022–4026. https://doi.org/10.1099/ijsem.0.001304
Author
Page authored by Henry Conaway, student of Prof. Jay Lennon at IndianaUniversity.