EBOV: Difference between revisions
No edit summary |
|||
| (88 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{ | {{Conway}} | ||
==Etiology== | [[Image: ebola4.jpeg |thumb|400px|right| The Ebola virus micrograph. From: http://www.thesundaytimes.co.uk/sto/culture/books/non_fiction/article1133688.ece]] | ||
==<b>Etiology</b>== | |||
===Taxonomy=== | ===Taxonomy=== | ||
| Order = [[Mononegavirales]] | |||
| Family = [[Filoviridae]] | |||
| Genus = [[Ebolavirus]] | |||
| Species = [[Zaire ebolavirus]] | | |||
===Description=== | ===Description=== | ||
The Ebola virus was first detected in 1976 in the Democratic Republic of the Congo near the Ebola river; hence, the virus was named after the location of origin. (CDC). | |||
It is a negative-sense, non-segmented, enveloped RNA virus belonging to the Filoviridae family. There are five identified species of the virus, each named after the region they were localized. The Zaire, Sudan, Ivory Coast, and Bundibugyo affect humans; whereas, the Reston virus only causes disease in non-human primates. The natural reservoir of the virus remains unknown. However, non-human primates and fruit bats, of <i>Hypsignathus monstrosus, Epomops franqueti and Myonycteris torquata </i> species, are considered zoonotic reservoirs of the virus. The Reston virus was isolated from cynomolgous monkeys imported from the Philippines to the United States and Italy. | |||
Infection is characterized by septic shock, hemorrhagic fever, coagulation of cells, and tissue necrosis. Some individuals are able to recover, but most do not survive. It remains unknown as to why some survive and others die. It is classified as a level 4 disease by the CDC. [[#References|[1]]] [[#References|[2]]] [[#References|[3]]] | |||
==Pathogenesis== | ==<b>Pathogenesis</b>== | ||
===Transmission=== | ===Transmission=== | ||
There are two types of exposures that contribute to acquiring the Ebola virus. The first, primary exposure, is due to travel in an ebola endemic area. The second mode of transmission is contact with bodily fluids of an infected host as well as utilizing unsterilized hospital equipment that came in contact with the virus. [[#References|[4]]] | |||
===Infectious dose and incubation=== | ===Infectious dose and incubation=== | ||
Upon invasion of host cells, the virus typically presents a 2-21 day incubation period. The infectious dose is very low; 1-10 aerosolized particles are sufficient to cause disease. [[#References|[2]]] | |||
===Epidemiology=== | ===Epidemiology=== | ||
As indicated by the chart provided by The World Health Organization, Ebola virus mainly affects regions in Africa. However, Reston strain did affect monkeys in the United States and Italy when they were imported from Italy. The Zaire virus outbreak seems to be the most predominant. The chart depicts outbreaks in Africa up until May 2012. [[#References|[1]]] | |||
[[Image: chart-1.jpeg |thumb|400px|center| Record of Ebola hemorrhagic outbreaks as of May 2012. From: http://www.who.int/mediacentre/factsheets/fs103/en/]] | |||
===Virulence Factors=== | ===Virulence Factors=== | ||
EBOV viral strand contains seven genes encompassed in a 19 kb genome. Gene products produced are a glycoprotein (GP) virion envelope, nucleoprotein (NP), nonstructural proteins VP30 and VP35, matrix proteins VP24 and VP40, and a viral polymerase. VP35 and the GP are the major contributors to the onset of disease. [[#References|[9]]] | |||
====VP35==== | ====VP35==== | ||
Previous studies demonstrate VP35 is protein virulence factor that serves as a cofactor for viral RNA polymerase complex, RNAi silencing suppressor and for viral assembly. In addition, VP35 promotes immune lapse by blocking antiviral signaling pathways and by denaturing the type I IFN system. Thus, the protein is able to replicate RNA and evade host cells. EBOV VP35 IID interferes with type I interferons (IFNs) and IFNα/β upon the engagement of PRRs and PAMPs, which in a normal immune response, produce the necessary type I IFNs. Transcription factors (IRF)3, IRF-7, and NKκ stimulate the IFNα/β to release IFNα and IFNβ. IFNα and IFNβ in turn stimulate antiviral activity genes like MHC class I and PKR. By activating these genes, we can reduce viral replication and in turn reduce the spread of infection. Inhibition of IFNα/β indirectly affects dendritic cell maturation because IFN production is reduced. [[#References|[7]]] [[#References|[9]]] [[#References|[11]]] | |||
The mechanism by which VP35 binds its RNA remains uncertain. However, structural studies of the protein reveal that it is comprised of two subdomains. The alpha subdomain is composed of a four helix bundle and the beta domain is composed of four antiparallel beta strands, alpha helix, and a polyproline helix. Furthermore, the VP35 contains two basic patches, one in the beta sheet subdomain and the other in the alpha helical subdomain. The beta sheet domain has Arg305, Arg12, and Lys309 that allow for further immune suppression. VP35 is also characterized by an “end cap” that is composed of hydrophobic Phe239 and Ile340; this hydrophobic area allows RNA to bind to VP35 IID; in turn, antagonize antiviral signaling pathways. The N-terminus of the protein is utilized to allow NP and VP40 binding, the formation of viral polymerase complex, PKR, and RNAi silencing suppression. Scientists believe that perhaps preventing the adherence of VP35, the ebola virus can be avoided in host cells. [[#References|[7]]] [[#References|[9]]] | |||
====GP==== | ====GP==== | ||
The glycoprotein (GP) envelope around EBOV allows the virus to adhere and fuse with host cells. Studies conducted in the past suggest that the GP holds a trimeric form with GP1 and GP2 (which are linked by a disulfide bond). A neutralizing antibody, KZ52, detected in a human EBOV survivor was utilized to crystalize the structure of the GP+antibody. GP1 attributes allow EBOV to attach to host cells. It is composed of a base, head, and glycan cap. The base forms a clamp that allows the stabilization of GP2 pre-fusion conformation. The GP2 subunit attributes allow EBOV to fuse with host cells. GP1 forms a chalice that is supported by GP2 whilst a glycan cap and mucin-like domain protect the receptor binding site. The neutralizing antibody detects proteins on the chalice base causing it to bind to GP1 via Van der Waals forces. EBOV then makes it’s entry into host cells via endocytosis. [[#References|[10]]] | |||
==<b>Clinical features</b>== | |||
===Symptoms=== | |||
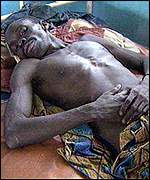
Ebola nfection is characterized by the onset of malaise, fever, myalgia, diarrhea, vomiting, and headaches. As the disease progresses, gastrointestinal bleeding, lymphopenia, neutrophilia, maculopapular rash, conjuctivitis, along with external bleeding may occur. Some patients are able to recover from the infection; however, it remains unknown as to why some recover and others die. Survivors are typically left with a range of maladies that include fatigue, bulimia, hearing loss, tinnitus, arthralgias, orchitis, and suppurative parotitis. [[#References|[4]]] [[#References|[9]]] | |||
[[Image: ebolavictim.jpeg |thumb|400px|right| Ebola patient. <br /> From: http://news.bbc.co.uk/2/hi/africa/567974.stm]] | |||
===Mortality=== | |||
Mortality rates are between 50-90%. It is possible to survive the Ebola virus; however, statistics suggest the EBOV is more fatal than the other species of the ebola virus. Epidemiological studies indicate that the Zaire species is predominant in infecting human hosts. [[#References|[9]]] | |||
==<b>Diagnosis</b>== | |||
Typically, clinical diagnosis can only be made after the first few days of symptoms because the early symptoms could have been caused by many other factors. | |||
Laboratory tests such as, enzyme-linked immunosorbent assay (ELISA), serum neturalization tests, antigen detection tests, virus isolation, and a reverse transcriptase polymerase chain reaction (RT-PCR) definitively allow the detection of the ebola virus in a patient. Immunohistochemistry testing and PCR can be conducted in patients that did not survive the disease. These tests are conducted under maximum biological containment conditions. [[#References|[1]]] [[#References|[2]]] [[#References|[3]]] | |||
==<b>Treatment</b>== | |||
Currently, vaccines and antivirals are not available to treat EBOV. Patients are provided supportive treatment that includes attention to: replacement of fluids, electrolytes, constant monitoring of blood pressure and oxygen levels, nutrition and comfort. [[#References|[1]]] | |||
==<b>Prevention</b>== | |||
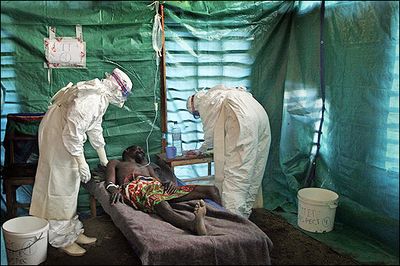
Infected individuals are quarantined in a facility that entails the necessary safety measures authored by the CDC and WHO. Health care professionals dress in biohazard suits before coming in contact with a patient. If biohazard suits are not available, protective clothing, goggles, gloves, and mask should be worn. To reduce the risk of acquiring the Ebola virus, avoid travelling to endemic areas in Africa. In addition, avoid consuming bush meats that are sold on street markets. Wear gloves, protective clothing, and a mask when caring for an ill that may possibly be affect with ebola. Also avoid coming in contact with an infected corpse. Wash hands frequently when travelling; if no soap is available, rub hands with 60% alcohol. Take careful precautions before coming in contact with the fluids of non-human primates from Africa or the Philippines. [[#References|[1]]] [[#References|[5]]] [[#References|[6]]] | |||
[[Image: ebov.jpeg |thumb|400px|left| Depiction of health care professionals dressed in biohazard suits. From: http://foxfromzim.files.wordpress.com/2010/05/ebola.jpeg]] | |||
==<b>Host Immune Response</b>== | |||
The lack of infected human tissue prevents the pathologic study of EBOV; thus, studies conducted in cynomolgus monkeys only suggest the possible mechanisms of infection in humans. | |||
The GP gives EBOV access to host cells by allowing the transfer of the viral genome into macrophages. Infected cells may trigger antigen-specific immune responses to prevent viral replications; however, if immune responses fail, death occurs 1-2 weeks after infection. The present GP allows the virus to conveniently adhere to cell surface lectins in addition to a variety of other molecules. ZEBOV impairs the function of macrophages and dendritic cells in the immune response system. These antigen presenting cells are able to trigger inflammation and coagulation; however, they lack the ability to prevent RNA replication in the host cells. | |||
RNA binds to PRRs on the macrophage surface to trigger cytoplasmic signaling to cause the migration of vasoactive molecules like cytokines (TNF-alpha and IL-1beta), chemokines (MIP-1alpha), and nitric oxide. The release of these molecules causes additional macrophages and monocytes to migrate to the infected cell. Neutrophils are then released to aid the present macrophages. Lipid mediators trigger the inflammation response, which causes vasodilation, increased endothelial permeability, and expression of adhesion molecules to trap leukoctyes. This allows the virus to skillfully invade host immune cells to cause hypotension and septic shock. In addition, the binding of coagulation factors to cells excites intracellular signaling pathways by phosphorylating cytoplasmic TF tails; this hinders macrophage function. | |||
Infected dendritic cells fail to express costimulatory molecules, MHC, and thus, prevent differentiation of lymphocytes. ZEBOV does not invade lymphocytes; however, the presence of the virus does initiate cell apoptosis causing lymphopenia and thus, septic shock. Natural killer cells, CD4 and CD8 cells are the prominent cell types affected during the course of the disease. Typically, these cells play a critical role in preventing viral replication. [[#References|[8]]] [[#References|[12]]] [[#References|[13]]] | |||
== | ==<b>References</b>== | ||
= | 1 [http://www.who.int/mediacentre/factsheets/fs103/en/ WHO Fact Sheet on Ebola haemorrhagic fever] | ||
<br> | |||
2 [http://www.phac-aspc.gc.ca/lab-bio/res/psds-ftss/ebola-eng.php Public Health Agency of Canada Ebola virus- Pathogen Safety Data Sheets] | |||
<br> | |||
3 [http://www.cdc.gov/ncidod/dvrd/spb/mnpages/dispages/ebola/qa.htm CDC Questions and Answers about Ebola Hemorrhagic fever] | |||
<br> | |||
4 [http://emedicine.medscape.com/article/216288-overview#aw2aab6b2b5 MedScape Ebola Virus] | |||
<br> | |||
5 [http://www.mayoclinic.com/health/ebolavirus/DS00996/DSECTION=tests%2Dand%2Ddiagnosis Mayo Clinic Ebola Virus and Marburg Virus] | |||
<br> | |||
6 [http://www.nlm.nih.gov/medlineplus/ency/article/001339.htm Medline Plus Ebola hemorrhagic fever] | |||
<br> | |||
7 [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3061251/ Daisy W Leung, Kathleen C Prins, Christopher F Basler, and Gaya K Amarasinghe. Ebolavirus VP35 is a multifunctional virulence factor. US National Library of Medicine and National Institute of Health] | |||
<br> | |||
8 [http://www.ncbi.nlm.nih.gov.ezproxy.lib.ou.edu/pubmed/15896665 Bray, Mike and Thomas W. Geisbert. Ebola virus: The role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. US National Library of Medicine and National Institute of Health] | |||
<br> | |||
9 [http://jvi.asm.org/content/77/18/9733 Sullivan, Nancy, Zhi-Yong Yang, and Gary J. Nabel. Ebola Virus Pathogenesis: Implications for Vaccines and Therapies. American Society for Microbiology] | |||
<br> | |||
10 [http://www.ncbi.nlm.nih.gov/pubmed/20198110 JE, Lee and Saphire EO. Ebolavirus glycoprotein structure and mechanism of entry. US National Library of Medicine and National Institutes of Health] | |||
<br> | |||
11 [http://www.ncbi.nlm.nih.gov/pubmed/17118636 Saito T, Gale M., Principles of intracellular viral recognition. US National Library of Medicine and National Institutes of Health] | |||
<br> | |||
12 [http://www.ncbi.nlm.nih.gov/pubmed/11352664 Gupta, M, S. Mahanty, R. Ahmed, and P.E. Rollin. Monocyte-derived human macrophages and peripheral blood mononuclear cells infected with ebola virus secrete MIP-1alpha and TNF-alpha and inhibit poly-IC-induced IFN-alpha in vitro. US National Library of Medicine and National Institutes of Health] | |||
<br> | |||
13 [http://www.ncbi.nlm.nih.gov/pubmed/12519925 Hotchkiss, R.S., and Karl I.E. The pathophysiology and treatment of sepsis. US National Library of Medicine and National Institutes of Health] | |||
<br> | |||
[[Image: OUA.png |thumb|400px|right| OU Microbiology in Arezzo, Italy]] | |||
[ | |||
Created by | Created by Bhumi Patel, student of Tyrrell Conway at the University of Oklahoma. | ||
Latest revision as of 14:41, 11 February 2016

Etiology
Taxonomy
| Order = Mononegavirales | Family = Filoviridae | Genus = Ebolavirus | Species = Zaire ebolavirus |
Description
The Ebola virus was first detected in 1976 in the Democratic Republic of the Congo near the Ebola river; hence, the virus was named after the location of origin. (CDC). It is a negative-sense, non-segmented, enveloped RNA virus belonging to the Filoviridae family. There are five identified species of the virus, each named after the region they were localized. The Zaire, Sudan, Ivory Coast, and Bundibugyo affect humans; whereas, the Reston virus only causes disease in non-human primates. The natural reservoir of the virus remains unknown. However, non-human primates and fruit bats, of Hypsignathus monstrosus, Epomops franqueti and Myonycteris torquata species, are considered zoonotic reservoirs of the virus. The Reston virus was isolated from cynomolgous monkeys imported from the Philippines to the United States and Italy. Infection is characterized by septic shock, hemorrhagic fever, coagulation of cells, and tissue necrosis. Some individuals are able to recover, but most do not survive. It remains unknown as to why some survive and others die. It is classified as a level 4 disease by the CDC. [1] [2] [3]
Pathogenesis
Transmission
There are two types of exposures that contribute to acquiring the Ebola virus. The first, primary exposure, is due to travel in an ebola endemic area. The second mode of transmission is contact with bodily fluids of an infected host as well as utilizing unsterilized hospital equipment that came in contact with the virus. [4]
Infectious dose and incubation
Upon invasion of host cells, the virus typically presents a 2-21 day incubation period. The infectious dose is very low; 1-10 aerosolized particles are sufficient to cause disease. [2]
Epidemiology
As indicated by the chart provided by The World Health Organization, Ebola virus mainly affects regions in Africa. However, Reston strain did affect monkeys in the United States and Italy when they were imported from Italy. The Zaire virus outbreak seems to be the most predominant. The chart depicts outbreaks in Africa up until May 2012. [1]

Virulence Factors
EBOV viral strand contains seven genes encompassed in a 19 kb genome. Gene products produced are a glycoprotein (GP) virion envelope, nucleoprotein (NP), nonstructural proteins VP30 and VP35, matrix proteins VP24 and VP40, and a viral polymerase. VP35 and the GP are the major contributors to the onset of disease. [9]
VP35
Previous studies demonstrate VP35 is protein virulence factor that serves as a cofactor for viral RNA polymerase complex, RNAi silencing suppressor and for viral assembly. In addition, VP35 promotes immune lapse by blocking antiviral signaling pathways and by denaturing the type I IFN system. Thus, the protein is able to replicate RNA and evade host cells. EBOV VP35 IID interferes with type I interferons (IFNs) and IFNα/β upon the engagement of PRRs and PAMPs, which in a normal immune response, produce the necessary type I IFNs. Transcription factors (IRF)3, IRF-7, and NKκ stimulate the IFNα/β to release IFNα and IFNβ. IFNα and IFNβ in turn stimulate antiviral activity genes like MHC class I and PKR. By activating these genes, we can reduce viral replication and in turn reduce the spread of infection. Inhibition of IFNα/β indirectly affects dendritic cell maturation because IFN production is reduced. [7] [9] [11]
The mechanism by which VP35 binds its RNA remains uncertain. However, structural studies of the protein reveal that it is comprised of two subdomains. The alpha subdomain is composed of a four helix bundle and the beta domain is composed of four antiparallel beta strands, alpha helix, and a polyproline helix. Furthermore, the VP35 contains two basic patches, one in the beta sheet subdomain and the other in the alpha helical subdomain. The beta sheet domain has Arg305, Arg12, and Lys309 that allow for further immune suppression. VP35 is also characterized by an “end cap” that is composed of hydrophobic Phe239 and Ile340; this hydrophobic area allows RNA to bind to VP35 IID; in turn, antagonize antiviral signaling pathways. The N-terminus of the protein is utilized to allow NP and VP40 binding, the formation of viral polymerase complex, PKR, and RNAi silencing suppression. Scientists believe that perhaps preventing the adherence of VP35, the ebola virus can be avoided in host cells. [7] [9]
GP
The glycoprotein (GP) envelope around EBOV allows the virus to adhere and fuse with host cells. Studies conducted in the past suggest that the GP holds a trimeric form with GP1 and GP2 (which are linked by a disulfide bond). A neutralizing antibody, KZ52, detected in a human EBOV survivor was utilized to crystalize the structure of the GP+antibody. GP1 attributes allow EBOV to attach to host cells. It is composed of a base, head, and glycan cap. The base forms a clamp that allows the stabilization of GP2 pre-fusion conformation. The GP2 subunit attributes allow EBOV to fuse with host cells. GP1 forms a chalice that is supported by GP2 whilst a glycan cap and mucin-like domain protect the receptor binding site. The neutralizing antibody detects proteins on the chalice base causing it to bind to GP1 via Van der Waals forces. EBOV then makes it’s entry into host cells via endocytosis. [10]
Clinical features
Symptoms
Ebola nfection is characterized by the onset of malaise, fever, myalgia, diarrhea, vomiting, and headaches. As the disease progresses, gastrointestinal bleeding, lymphopenia, neutrophilia, maculopapular rash, conjuctivitis, along with external bleeding may occur. Some patients are able to recover from the infection; however, it remains unknown as to why some recover and others die. Survivors are typically left with a range of maladies that include fatigue, bulimia, hearing loss, tinnitus, arthralgias, orchitis, and suppurative parotitis. [4] [9]
Mortality
Mortality rates are between 50-90%. It is possible to survive the Ebola virus; however, statistics suggest the EBOV is more fatal than the other species of the ebola virus. Epidemiological studies indicate that the Zaire species is predominant in infecting human hosts. [9]
Diagnosis
Typically, clinical diagnosis can only be made after the first few days of symptoms because the early symptoms could have been caused by many other factors. Laboratory tests such as, enzyme-linked immunosorbent assay (ELISA), serum neturalization tests, antigen detection tests, virus isolation, and a reverse transcriptase polymerase chain reaction (RT-PCR) definitively allow the detection of the ebola virus in a patient. Immunohistochemistry testing and PCR can be conducted in patients that did not survive the disease. These tests are conducted under maximum biological containment conditions. [1] [2] [3]
Treatment
Currently, vaccines and antivirals are not available to treat EBOV. Patients are provided supportive treatment that includes attention to: replacement of fluids, electrolytes, constant monitoring of blood pressure and oxygen levels, nutrition and comfort. [1]
Prevention
Infected individuals are quarantined in a facility that entails the necessary safety measures authored by the CDC and WHO. Health care professionals dress in biohazard suits before coming in contact with a patient. If biohazard suits are not available, protective clothing, goggles, gloves, and mask should be worn. To reduce the risk of acquiring the Ebola virus, avoid travelling to endemic areas in Africa. In addition, avoid consuming bush meats that are sold on street markets. Wear gloves, protective clothing, and a mask when caring for an ill that may possibly be affect with ebola. Also avoid coming in contact with an infected corpse. Wash hands frequently when travelling; if no soap is available, rub hands with 60% alcohol. Take careful precautions before coming in contact with the fluids of non-human primates from Africa or the Philippines. [1] [5] [6]
Host Immune Response
The lack of infected human tissue prevents the pathologic study of EBOV; thus, studies conducted in cynomolgus monkeys only suggest the possible mechanisms of infection in humans.
The GP gives EBOV access to host cells by allowing the transfer of the viral genome into macrophages. Infected cells may trigger antigen-specific immune responses to prevent viral replications; however, if immune responses fail, death occurs 1-2 weeks after infection. The present GP allows the virus to conveniently adhere to cell surface lectins in addition to a variety of other molecules. ZEBOV impairs the function of macrophages and dendritic cells in the immune response system. These antigen presenting cells are able to trigger inflammation and coagulation; however, they lack the ability to prevent RNA replication in the host cells.
RNA binds to PRRs on the macrophage surface to trigger cytoplasmic signaling to cause the migration of vasoactive molecules like cytokines (TNF-alpha and IL-1beta), chemokines (MIP-1alpha), and nitric oxide. The release of these molecules causes additional macrophages and monocytes to migrate to the infected cell. Neutrophils are then released to aid the present macrophages. Lipid mediators trigger the inflammation response, which causes vasodilation, increased endothelial permeability, and expression of adhesion molecules to trap leukoctyes. This allows the virus to skillfully invade host immune cells to cause hypotension and septic shock. In addition, the binding of coagulation factors to cells excites intracellular signaling pathways by phosphorylating cytoplasmic TF tails; this hinders macrophage function.
Infected dendritic cells fail to express costimulatory molecules, MHC, and thus, prevent differentiation of lymphocytes. ZEBOV does not invade lymphocytes; however, the presence of the virus does initiate cell apoptosis causing lymphopenia and thus, septic shock. Natural killer cells, CD4 and CD8 cells are the prominent cell types affected during the course of the disease. Typically, these cells play a critical role in preventing viral replication. [8] [12] [13]
References
1 WHO Fact Sheet on Ebola haemorrhagic fever
2 Public Health Agency of Canada Ebola virus- Pathogen Safety Data Sheets
3 CDC Questions and Answers about Ebola Hemorrhagic fever
4 MedScape Ebola Virus
5 Mayo Clinic Ebola Virus and Marburg Virus
6 Medline Plus Ebola hemorrhagic fever
7 Daisy W Leung, Kathleen C Prins, Christopher F Basler, and Gaya K Amarasinghe. Ebolavirus VP35 is a multifunctional virulence factor. US National Library of Medicine and National Institute of Health
8 Bray, Mike and Thomas W. Geisbert. Ebola virus: The role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. US National Library of Medicine and National Institute of Health
9 Sullivan, Nancy, Zhi-Yong Yang, and Gary J. Nabel. Ebola Virus Pathogenesis: Implications for Vaccines and Therapies. American Society for Microbiology
10 JE, Lee and Saphire EO. Ebolavirus glycoprotein structure and mechanism of entry. US National Library of Medicine and National Institutes of Health
11 Saito T, Gale M., Principles of intracellular viral recognition. US National Library of Medicine and National Institutes of Health
12 Gupta, M, S. Mahanty, R. Ahmed, and P.E. Rollin. Monocyte-derived human macrophages and peripheral blood mononuclear cells infected with ebola virus secrete MIP-1alpha and TNF-alpha and inhibit poly-IC-induced IFN-alpha in vitro. US National Library of Medicine and National Institutes of Health
13 Hotchkiss, R.S., and Karl I.E. The pathophysiology and treatment of sepsis. US National Library of Medicine and National Institutes of Health
Created by Bhumi Patel, student of Tyrrell Conway at the University of Oklahoma.