Encephalitozoon hellem: Difference between revisions
| Line 23: | Line 23: | ||
==Genome structure== | ==Genome structure== | ||
<i>Encephalitozoon</i> The Genome size of is approximately 2.39 Mb ([http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=11456324&ordinalpos=47&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum=abstract 7]). Analysis of the Internal Transcribed Spacer (ITS) sequence of the rRNA genes and SSU rRNA sequences of various isolates indentified 3 types of genotypes for <i>E. hellem</i>, 1A, 1B, and 1C. Each of these genotypes have 1253 bp, 1313 bp, and 1373 bp respectively and all have identical ITS sequences. While ITS sequences served as a valuable marker in researching <i>E. hellem</i>, the polar tubule protein (PTP) gene proved to be more valuable as it found an additional genotype, 2B, having 1421 base pairs ([http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=88110=abstract 5]). Genotype 2B has very different ITS and SSU rRNA sequences and extensive differences in PTP sequences. Later studies revealed two more genotypes, 2A and 2B. While these six different genotypes have been discovered the significance of <i>E. hellem's</i> genetic diversity is unclear and the number of <i>E. hellem</i> isolates is very limited leaving researchers without enough information to compare genotype distribution among humans and other hosts such as birds ([http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=88110=abstract 5]). | <i>Encephalitozoon</i> species have the smallest genomes reported to date among all single-celled eukaryotes. It is suggested that this is because of an early divergence of microsporidia that are no longer supported by current phylogenic data. The Genome size of is approximately 2.39 Mb ([http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=11456324&ordinalpos=47&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum=abstract 7]). Analysis of the Internal Transcribed Spacer (ITS) sequence of the rRNA genes and SSU rRNA sequences of various isolates indentified 3 types of genotypes for <i>E. hellem</i>, 1A, 1B, and 1C. Each of these genotypes have 1253 bp, 1313 bp, and 1373 bp respectively and all have identical ITS sequences. While ITS sequences served as a valuable marker in researching <i>E. hellem</i>, the polar tubule protein (PTP) gene proved to be more valuable as it found an additional genotype, 2B, having 1421 base pairs ([http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=88110=abstract 5]). Genotype 2B has very different ITS and SSU rRNA sequences and extensive differences in PTP sequences. Later studies revealed two more genotypes, 2A and 2B. While these six different genotypes have been discovered the significance of <i>E. hellem's</i> genetic diversity is unclear and the number of <i>E. hellem</i> isolates is very limited leaving researchers without enough information to compare genotype distribution among humans and other hosts such as birds ([http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=88110=abstract 5]). | ||
Pulsed-field gel electrophoresis (PFGE) is most often used for typing <i>Encephalitozoon hellem</i> and has found two karyotypes A and B with 7 and 8 chromosomal bands respectively. | Pulsed-field gel electrophoresis (PFGE) is most often used for typing <i>Encephalitozoon hellem</i> and has found two karyotypes A and B with 7 and 8 chromosomal bands respectively. | ||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
Revision as of 02:51, 27 August 2007
A Microbial Biorealm page on the genus Encephalitozoon hellem
Classification
Higher order taxa
Superkingdom, Kingdom, Phylum, Suborder, Family, Genus:
Eukaryota, Fungi, Microsporidia, Apansporoblastina, Unikaryonidae, Encephalitozoon
Species
Encephalitozoon hellem
Description and significance
Encephalitozoon hellem was initially dectected and isolated in 1991 from three AIDS patients who were suffering from keratoconjunctivitis among other ailments. Conjunctival scrapings and corneal tissue samples were taken from each of the three patients and were tested using SDS-PAGE analysis and Western Blotting (4). All three isolates had identical banding patterns and appeared similar to another species of Encephalitozoon, E. cuniculi, and the new AIDS related microsporidian was then given its name, Encephaliozoon hellem.
In 1994, another AIDS patient experiencing a foreign body sensation in his left eye had a conjunctival swab tested using a calcofluor stain and a fluorescent-antibody stain with murein antiserum raised against E. hellem. A cross-reaction occured with another species of Encephalitozoon, so a PCR test was performed. It was found that the amplified rRNA generated digestion patterns by restriction endonuclease FokI that were indentical to the digestion patterns of E. hellem (3). These observations allowed researchers to conclude that Encephalitozoon hellem causes keratoconjunctivitis in addition to other disseminated infections in immunosuppressed patients. Doctors and researchers soon found that patients treated with albendazole and topical fumagillin responded rapidly with no opthalmologic signs (3).
Encephalitozoon hellem is a unicellular microsporidian that is though to be fecal borne. It is a spore forming parasite whose spores measure approximately 1x1.5-2.0 microns in diameter (4). E. hellem has been named one of the four most common human microsporidian parasites, with its human hosts consisting mainly of immunodificient patients. The genotyping of this microsporidian has been helpful in understanding other forms of human microsporidosis and their transmission (5). Microsporidia such as E. hellem is culturable from affected tissues and useful for species-species diagnosis. While culturing of these microbes are time-consuming and requires expert skill, species-species identification of microsporidia is important because of various levels of drug responses of the differing species (3).
Genome structure
Encephalitozoon species have the smallest genomes reported to date among all single-celled eukaryotes. It is suggested that this is because of an early divergence of microsporidia that are no longer supported by current phylogenic data. The Genome size of is approximately 2.39 Mb (7). Analysis of the Internal Transcribed Spacer (ITS) sequence of the rRNA genes and SSU rRNA sequences of various isolates indentified 3 types of genotypes for E. hellem, 1A, 1B, and 1C. Each of these genotypes have 1253 bp, 1313 bp, and 1373 bp respectively and all have identical ITS sequences. While ITS sequences served as a valuable marker in researching E. hellem, the polar tubule protein (PTP) gene proved to be more valuable as it found an additional genotype, 2B, having 1421 base pairs (5). Genotype 2B has very different ITS and SSU rRNA sequences and extensive differences in PTP sequences. Later studies revealed two more genotypes, 2A and 2B. While these six different genotypes have been discovered the significance of E. hellem's genetic diversity is unclear and the number of E. hellem isolates is very limited leaving researchers without enough information to compare genotype distribution among humans and other hosts such as birds (5).
Pulsed-field gel electrophoresis (PFGE) is most often used for typing Encephalitozoon hellem and has found two karyotypes A and B with 7 and 8 chromosomal bands respectively.
Cell structure and metabolism
Describe any interesting features and/or cell structures; how it gains energy; what important molecules it produces. Encephalitozoon hellem is a unicellular, intracellular microsporidian species (1)
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
E. hellem was first isloated in HIV patients, but has also been found to infect mice, birds, and even bats. When isolating E. hellem in the HIV patient, there were 24 monoclonal antibodies that were used against not only E. hellem, but also two other species of Encephalitozoon, E. intestinalis and E. cuniculi. The antibodies did not react with either of these two, indicating antibody specificity across the three species. Antigenic diversity of the different karyotypes of E. hellem was demonstrated as well when two monoclonal antibodies reacted with one karyotype B and not karyotype A (1).
Experiments exposing chickens to E. hellem found that the microbe was detectable in the feces of the animals for up to 19 days, showing that E. hellem is potentially fecal borne and that chickens are a very likely host. While a definite route of infection is not well documented, it has been assumed that transmission of E. hellem most likely occurs by direct contact or by the ingestion of contaminated food or water (2).
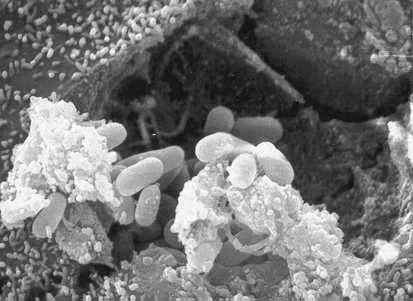
Once a host has been found, E. hellem like other microsporidia are spore forming parasites that use their unique polar tubule as a means of infecting their hosts. A mature, resistant spore will extrude the polar tubule and through this, the sporoplasm will be injected into the host cell. The sporoplasm is then able to multiply inside of the host cell via binary fission or multiple fission. Like the photo above illustrates, the spores will continue to multiply until the host cell cytoplasm is filled, causing it to burst and release more spores into the surroundings that will mature and carry on the infection to other cells (6).
The clinical manifestations of E. hellem include keratoconjunctivitis, infection of respiratory and genitourinary tract, and disseminated infection. With E. hellem being one of the four most common human microsporidian parasites, hosts are usually HIV patients but infections have also been reported to infect immunodificient patients that do not have HIV (6).
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
Enter summaries of the most recent research here--at least three required
References
1. Mo L., Drancourt M. Antigenic diversity of Encephalitozoon hellem demonstrated by subspecies-specific monoclonal antibodiesJ. Eukaryotic Microbiology 49, 249-54 (2002)
2. Fayer, R. et. al Detection of Encephalitozoon Hellem in Feces of Experimentally Infected ChickensJ. Eukaryotic Microbiology 50, 5743-575 (2003)
3. Didier, E.S. et. al Diagnosis of disseminated microsporidian Encephalitozoon hellem infection by PCR-Southern analysis and successful treatment with albendazole and fumagillinJ Clinical Microbiology, 34, 947–952 (1996)
4. Didier, E.S. et. al Isolation and characterization of a new human microsporidian, Encephalitozoon hellem (n. sp.), from three AIDS patients with keratoconjunctivitisJ Infect. Dis. 163,617-21 (1991)
5. Xiao, L. et. al Genotyping Encephalitozoon hellem Isolates by Analysis of the Polar Tube Protein GeneJ Clinical Microbiology 39, 2191–2196 (2001)
6. Microsporidiosis Centers for Disease Control & Prevention National Center for Infectious Diseases Division of Parasitic Diseases
7. Delarbre, S. et. al Genetic diversity in the microsporidian Encephalitozoon hellem demonstrated by pulsed-field gel electrophoresis J. Eukaryotic Microbiology 48, 471-474 (2001)
Edited by Tiffany Myer, student of Rachel Larsen
