Epstein-Barr virus (EBV) and EBV-associated lymphomas
By Yangyang Liu (Kenyon '23)
Introduction

Epstein-Barr virus (EBV), formally called Human gammaherpesvirus 4, are a group within the Lymphocryptovirus genus of the Herpesviridae family. EBVise double-stranded DNA viruses that use RNA polymerase for mRNA synthesis based on their negative-strand as the template. EBV Infection occurs through oral transfer of saliva as well as genital secretions and more than 90% of normal adults would gain adaptive immunity after their primary EBV infection. The remaining EBV virus would remain in an asymptomatic latent state for a lifetime within resting B-cells, and a healthy adult with a working immune system would be able to contain the infection with the help of their cytotoxic T cells (CTLs), lymphocyte CD8+ and CD4+, and natural killer (NK) cells [1]. Only a small subset encountering life-threatening diseases due to them unable in maintaining the virus within the latent state. In an uncontrolled situation, EBV-driven lymphoproliferative disorders and lymphomas could develop with the patients. EBV-associated cancers are a common example among the ~15% of all human cancers involving a virus infection [2].
Some of the most well-known illnesses that are caused by EBV are mononucleosis, Burkitt’s lymphoma, Hodgkin’s lymphoma, non-Hodgkin lymphoma, post-transplant lymphoproliferative disease (PTLD), nasopharyngeal carcinoma, and numerous other types of cancer [3]. So far, there is limited success in antivirals development to combat EBV infection. With EBV’s putative role in carcinogenesis, it is important to learn about the mechanisms by which the virus alters host cell characteristics and thus evades the immune system of the hosts. This page investigates how different latency patterns of EBV infection could lead to numerous types of lymphoma, the role of EBV in rare EBV-associated NK-cell lymphoproliferative diseases, as well as possible treatment for EBV-T/NK-LPDs.
Overall paper length should be 3,000 words, with at least 3 figures. Include some current research in each topic, with at least one figure showing data per section.
Identifying Latency Patterns of EBV Infection
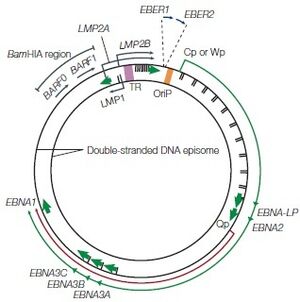
During primary in vivo infection, EBV preferentially infects B-lymphocytes by binding to the CD21 receptor on the cell surface, it will also bind to HLA class II molecules as a co-receptor. In most of the patients, the EBV episome would remain in the latent cycle in resting memory B cells after the primary infection while developing strategies to alter the pattern of the host’s cytotoxic T-lymphocyte (CTL) gene expression. Although there are nearly 100 viral proteins in a regular EBV genome, after infection, the infected resting memory B cells will limit the gene expression to nine viral latent proteins to evade immune recognition. The nine viral latent proteins are composed of six nuclear antigens EBNAs-1, -2, -3a, -3b, -3c, and -LP as well as three latent membrane proteins LMP-1, -2a, -2b. 2 small non-coding RNAs, EBER-1, EBER-2, and BamHI-A rightward transcripts (BART) are also expressed. The expression of nuclear antigens maintains the viral genome, they are also responsible for controlling the expression patterns of the three latent membrane proteins. Different expression combinations of EBNAs and LMPs can lead to different EBV latency programs and the associated lymphomas [4]. Type I latency is only seen in Burkitt’s lymphoma (BL) and selectively express EBNA-1. Type II latency expresses EBNA-1 along with LMP-1 and LMP-2, the common lymphoma for type II latency to be found are Hodgkin’s lymphoma (HL) and peripheral T-cell lymphoma (PTCL). The third and last EBV latency program type III, is commonly found in PTLD and AIDS-related lymphoma. All nine EBV latent proteins are expressed in type III latency [4].
MiRNAs for Viral Latency Identification

The EBV latency type classification is still ongoing work, as the traditional classification system stated above is only based on a few molecular markers of EBV latency which introduces limitations to typing of EBV latency types. As different latent infections are associated with different tumors, inaccurate latency identification is an academic and clinical issue.
MicroRNAs (MiRNAs) species have been found in EBV-infected cells, and a total of 44 EBV miRNA have been reported to be involved in EBV viral latency and tumorigenesis. Viral miRNAs are usually hard-to-detect and are 19-23 nt in length [5]. They are crucial in maintaining persistent EBV viral infections by promoting tumorigenesis in hosts and down-regulating viral early gene expression. Furthermore, the viral miRNAs are also able to regulate host gene expression at the post-transcriptional level to assist the virus in evading host immune responses. EBV was first found in 2004 to be encoding viral miRNAs, and the to-date 44 mature EBV miRNAs are produced and expressed as two independent transcripts from BHRF1 and BARTs intron prior to splicing [5]. Those abundant markers are a hopeful option to be turned into biomarkers for EBV latency types identification in the future.
In a recent study, highly sensitive and specific quantitative real-time RT-PCR (qRT-PCR) had been combined with microarray assay for the analysis of patterns of EBV miRNA transcriptomes that are associated with EBV patency types in tumor cells.
EBV-associated Rare NK-cell Lymphoproliferative Diseases
Treatment for EBV-T/NK-LPDs
Conclusion
References
- ↑ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3305910/#:~:text=Epstein%2DBarr%20virus%20(EBV),driven%20lymphoproliferative%20disorders%20and%20lymphomas
- ↑ https://onlinelibrary.wiley.com/doi/abs/10.1002/rmv.456
- ↑ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3415781/
- ↑ 4.0 4.1 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3305910/
- ↑ 5.0 5.1 https://virologyj.biomedcentral.com/articles/10.1186/1743-422X-10-314
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2021, Kenyon College.
