Feline Immunodeficiency virus
By: Emily Nutt
Introduction
Approximately 1.5 to 3 percent of all healthy cats in the United States are infected with Feline Immunodeficiency Virus or FIV. Sick cats are even more likely to contract FIV since they have a 15 percent chance of contracting the virus [2]. FIV is a type of lentivirus, a retrovirus that propagates at a slow pace. This slow propogation causes a huge gap of months to years from when the cat becomes infected with the virus and when it begins to show symptoms. A more widely known lentivirus is that of Human Immunodeficiency Virus or HIV. Unlike HIV, FIV infects cats through biting that occurs during fighting. HIV and FIV infect their host T-cells in a similar manor because they are both retroviruses.
All retroviruses use reverse transcriptase to covert their RNA genome into double-stranded DNA, which is then transcribed and translated to create new virions. These types of viruses carry the protein reverse transcriptase in side the viron instead of an early gene being translated to form the protein. In fact the reverse transcriptase is already bound to the viral RNA with a primer attached before the virus infects the host cell [7]. Like all viruses, retroviruses bind a receptor protein on the host cell and fuse with the host membrane. Once the capsid has been uncoated the RNA is converted to double-stranded DNA through the reverse transcriptase. The new DNA copy then enters the nucleus and integrates into the host genome. At first the viral genome is able to replicate slowly only creating a few virions and thus not directly affecting the host. The host cell’s RNA polymerase and ribosomes create virions through transcription and translation of viral genes. The viruses are assembled and then bud out of the host cell to infect new host cells [7]. After sometime the host cell can then generate a large amount of virions, thus causing the host to show symptoms of immunodeficiency. However it is unknown how this process of rapid viral growth occurs and why viruses like FIV can infect the host for months to years without the host showing symptoms.
The structure and process of lentivirus infections characterizes the three stages of FIV infection. The first stage of FIV is during the first two to four months after infection when the infected cat shows signs of short term illness. The infected cat may show symptoms like increased body temperature, and enlarged lymph nodes [1]. This short term illness could be a result of early and sparse virion replication in the host. In the second stage of FIV, the cat appears completely healthy with no signs of illness. However in the third phase of the illness the cat expresses signs of a compromised immunsystem. Signs of immunodeficiency include; weight loss, inappetence, high body temperature, gingivitis, infected lymphnodes, and abnormal cell growth resulting in tumors [1]. The three stages of FIV are similar to that of the stages of HIV, both stages of infection are characterized by the retrovirus lifecycle. The genome of FIV reveals important viral proteins and thus potential vaccine targets, as well as showing how the virus evolved.. The evolution of the virus explains how both domestic cats and exotic or non-domestic cats were initially infected with FIV. The viral evolution also reveals how the virus has mutated to become specialized for each species of cat. Further analysis of the genome of FIV and how it relates to the evolution of the virus is described below.
The Genome of FIV
All lentiviruses are made up of several key genes; gag, pol, env, rev, orfA and vif. Gag and Gag-Pol create the core of the FIV virion. The gag protein is modified post-transcription to create proteins for the viral matrix, capsid, and nucleocapsid. The pol gene produces a protease, reverse transcriptase, integrase and a protease-like protein. The protease-like protein is not found in primate lentiviruses. However it is found in FIV and maybe used to prevent misincorporation of the viral genome. Other evidence suggests that the protease like-protein is needed for efficient growth of infected T lymphocytes [8]. The vif protein is conserved in both HIV and FIV, and plays a role in the latency of FIV infection in cats. The rev protein functions as a stability and transport of unspliced viral RNA molecules to the nucleus of the host cell. Open reading frame A or orf A is induces host cell arrest at the second gap phase of the cell cylce [3]. Only gag and pol are the genes are in the primary viral genome and it is only after splicing that the other viral genes are expressed.
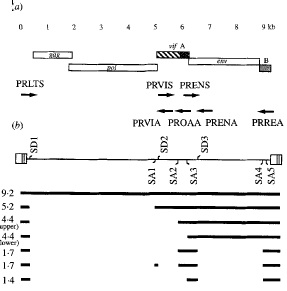
Splicing plays a key role in the expression of env, rev, orfA and vif genes of FIV. The vif and env genes are created through a single splicing of the primary transcript, while the rev gene arises from multiple splicing events. Figure 2 reveals the different splice sites found for the FIV genome. Part (b) of Figure 2 shows the different splice donor sites (SD) and the splice acceptor sites (SA) as well as the spliced mRNA produced from the genome [8]. In Figure 2 the numbers by the spliced products reveal the kilo bases of each RNA product. The 4.4 kb mRNA contains both the orfA and env open reading frame. The larger mRNA contains the complete orfA gene [8]. I n Figure 2 there are two 4.4 kilo base mRNAs both produce env and orf genes, however the 4.4 kilo bases with a longer orfA mRNA contains the whole gene of orfA. The 1.4 kb mRNA contains the three exons of the rev gene. The mRNA with 5.2 kb contains the vif gene in the larger mRNA product. The 1.7 kb mRNA contains pieces of both orfA and orfB, however the exact products these mRNAs produce is uncertain [8]. Splicing of the viral genome allows for FIV to carry a smaller genome that produces many different genes. The gag and pol genes remain quite conserved throughout different FIV strains, however the other genes vary greatly between different feline immunodeficiency viruses. This difference in env, rev, vif and orfA between types of FIV is due to different splice sites on the viral genome.
Genomic Variation of FIV
FIV is host specific making small mutations to evolve for each species of cat it infects. These mutations are typically occur in genes other than the pol gene. This gene is highly conserved in all lentiviruses because it contains the gene for reverse transcriptase a protein unique to all retroviruses [4]. Divergence in FIV genes changes the viral proteins, structure and function thus allowing the virus to evolve to be species specific.
Genomes of various FIV strains have been sequenced from the Felidae family and these FIV genomes are labeled by the species they infect. Several Felidae species are Puma concolor (cougar), Panthera leo (African Lion), Otocolobus manul (Pallas cat), and Felis catus ( domestic cat). The strains of FIV that infect each species are labeled FIV and then the first letter of the genus and the first two letters of the species name that the virus infects. Thus FIV-Fca is the strain of FIV that infects Felis catus. Figure 3 reveals the gene divergence of Puma concolor, Otocolobus manul, and Felis catus from Panthera leo[6]. The numbers in Figure 3 represent the percent divergence from FIV-Ple. In general Figure 3 shows that there is more divergence in env, vif, and orfA genes of FIV-Ple from the other cat species.
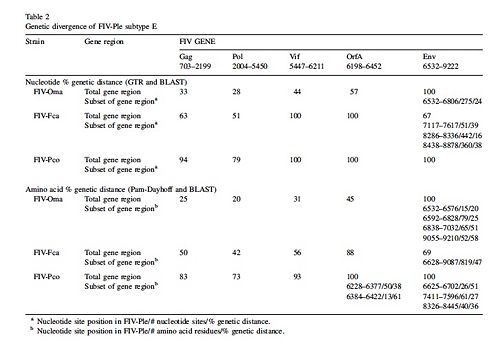
Figure 3 reveals that FIV-Ple genome has the least divergence from FIV-Oma genome, followed by FIV-Fca and the most divergence from FIV-Pco [6]. The similarities between FIV-Ple and FIV-Oma show that the two strains of the virus evolved from a common ancestor. Compared to the FIV-Oma genome, the pol gene of FIV-Ple is most related to the pol gene of FIV-Oma. The env gene of FIV-Ple is least related to the env gene of FIV-Oma. The next closest relative of FIV-Ple is FIV-Fca. Despite the large genetic divergence between env of FIV-Fca and env in FIV-Ple, both env genes share two common sequences [6]. Two regions of env gene in FIV-Ple are very similar to the same regions found in the env gene of FIV-Fca. The only part of the env gene that is similar to FIV-Oma is the exon used for the rev gene. This variation of the env gene reveals that recombination with FIV-Fca may have occurred. Recombination may also have occurred with another strain of FIV that has yet to be sequenced [6]. Scientists believe that a recombination event occurred in the env gene of FIV-Ple because it is most divergent from the genome of FIV-Oma. Since FIV-Oma and FIV-Ple are so similar it is assumed they diverged from the same ancestor and thus should have more similar env genomes. However since the two strains do not have similar genes, then recombination during the evolution of FIV-Ple seems most reasonable explanation for this difference[6]. Despite the differences in the env gene of FIV-Ple overall the viral genome is most closely related to FIV-Oma.
The gene variation of different strains of FIV reveals that the virus must evolve to fit each species of cat. Although only several viral genomes of several cat species have been completely sequenced, it is clear that each strain of FIV is species specific. This evolution of FIV to fit each particular species of cat is reasonable because each species of cat is different. For example each species of cat may have different T-cell receptors and thus the virus would have to mutate to bind to the new T-cell receptors [9]. It has also been shown that the feline immunsystem produces viral restriction factors that will limit unadapted viruses. Thus each strain of FIV must mutate to avoid the viral restriction factors of each feline species. The strains of FIV would also have to mutate to avoid each species conventional immune responses. The immune system of each species would vary because most species live in different environments and are thus affected by different pathogens. Differences in feline immune responses, restriction factors and T-cell receptors would cause each strain of FIV to mutate so that the virus could successfully infect each feline species.
The genomic variation of each strain of FIV not only reveals how FIV is able to mutate to avoid different immune systems of feline species, but it also reveals the evolution of FIV throughout felines in particular the Felidae. The Felidae is a family of cats that includes domestic cats, lions, leopards, cheetahs, hyaenidae, and cougars.
Evolution of feline immunodeficiency virus
FIV is found in feline species throughout the world including: Puma concolor in North and South America, Panthera leo in Africa, Felis catus, Otocolobus manul in Asia, Panther pardus (leopard) in Africa, Acinonyx jubatus (cheetah) and Crocuta corcuta (Hyena) in Africa. The phylogenetic tree reveals the complex evolution of FIV in different species of the Felidae family. FIV in Puma concolor have long branching revealing that the viral divergence occurred quite a long time ago [6]. FIV in Panthera leo has the most variation revealing that the virus probably began in Africa. Felis catus FIV diverged much later in history and seems to be a different variation of FIV from any other type of FIV in the Felidae family [6]. Some strains of FIV appear to be transmitted interspecies, however for this to occur serious obstacles must be overcome. Overall it appears that FIV first began in lions and spread throughout the world millions of years ago.
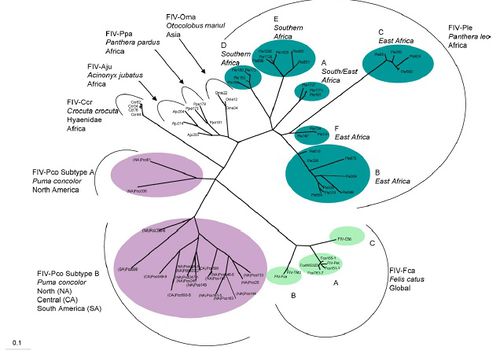
Out of all the species in Felidae FIV in Panthera leo has the most variation. There are six different varieties of FIV-Ple. FIV-Ple is divided geographically among Eastern and Southern Africa, however it appears FIV-Ple has more variation in Eastern Africa than in southern. The other reason FIV-Ple maybe so variable is because of the social practices of lions compared to other domestic cats [6]. Lions live in prides instead of living solitarily like cheetahs or cougars. Also lions incorporate biting and fighting in their everyday lifestyle, for example male lions are constantly taking over new prides. The geographical genetic variation is most extensive in FIV-Ple than other species of Felidae.
Puma concolor FIV is less variable because the species of cat is less social, however despite the lack of variability the strain of FIV appears to be very old. The age of FIV-Pco is seen in the long branching of the phylogenetic tree [6]. There are only two distinct variations of FIV-Pco and the two branches are more related to one another than other FIV strains in Felidae. These two strains have been in both species of cougar for a long time and seem to not cluster due to geographic location like other FIV in cats [6]. The two strains of FIV-Pco have long branches from their common ancestor showing that they are quite divergent between each other. However both FIV-Pco are more closely related to one another than to FIV in other Felidae species. The first group of FIV-Pco, FIV-Pco A contains two different strains of FIV located in North America. These stains are highly divergent from the other FlV-Pco B. FIV-Pco B is composed of different variation of the virus found in North, Central and South America. It is believed that FIV-Pco was first present in South American Puma concolor, which then re-colonized North America. Once the pumas were in North America they evolved and so did the virus to yield a slightly diverged version from the South American strain. These strains do not cluster geographically within the North American and South American strains thus revealing that South American pumas already had FIV when they re-colonized North America [6]
Overall the FIV-Pco is grouped less distinctly on geographical location than FIV-Ple. FIV-Ple has different strains for lions in different locations of East and South Africa. However FIV-Pco has two distinct strains one that is just found in North America and one strain that is found in North, Central and South America. This difference in grouping for FIV-Ple and FIV-Pco may be the difference of revealing that FIV-Ple is older than FIV-Pco [6]. However it is still unknown why FIV clusters more geographically for Panthera leo than in Puma concolor.
FIV in the domestic cat seems to have evolved more recently than both FIV-Ple and FIV-Pco. FIV-Fce to be greatly divergent from all other strains of FIV. Domestic cats evolved much later than lions and pumas, thus FIV-Fca has also evolved much later than FIV-Ple and FIV-Pco [6]. Also FIV-Fca greatly diverged from the other species of FIV that infect other cat species in the family of Felidae. This great divergence may be due to the fact that none of the close relatives to the domestic have been infected with FIV. It is still unknown how domestic cats contracted FIV when none of its close relatives or ancestors test positive for FIV. However this figure does reveal that FIV began in lions and diverged into other species through the evolution of cats in Felidae family.
The phylogenetic tree also shows some small evidence for FIV transmission between species in the Felidae family, in particular between Panthera pardus and Acinonyx jubatus. Both theses species of felines live in the National Park in Tanzania and the FIV that infects both species is very closely related. However in the evolution of felines Acinonyx jubatus is more closely related to Puma concolor than to Panthera pardus [6]. No other two species of felines that are that divergent are infected with very undivergent FIV strains. If interspecies transmission did occur between the leopard and the cheetah, FIV would have to over come some serious barriers [6]. The barriers the virus would have to over come include different T-cell receptors, variation in cell restriction proteins and the animals’ immune response [9]. For example some restrictive proteins cause cytosine deamination thus preventing complete transcription of the viral genome. However different species of felines may have restrictive proteins and a different immune response. These different responses would require the virus to mutate efficiently to effectively infect a new species of felines. Therefore this similarity in FIV between Panthera pardus and Acinonyx jubatus could be due to other means including geographic location.
Overall it appears that FIV infected Panthera leo first and then spread throughout the world as lions moved from Africa to Asia and the Americas in the late Pliocene age [6]. However it appears that behavioral and geographical barriers effected the evolution of FIV within each feline species. Also it is still unknown how domestic cats contracted FIV since none of their close relatives test positive for FIV. Also FIV in domestic cats appears most divergent from all other species in the Felidae species [6].
Conclusion
The genome of FIV reveals that different splicing sites would lead to variation in the expression of the FIV genome, thus allowing the virus to effectively infect multiple species of cats. The evolution of FIV in the Felidae family does not follow the exact evolution of the felines in family of cats. However, FIV evolution is affected by the behavior and geographic location of the felines in which it infects. By understanding the genome, genomic variation and evolution of FIV scientists can begin to understand HIV.
Like HIV, FIV can infects T-cells and causes immunodeficiency in its hosts. Also FIV has a similar genome to FIV especially in the pol gene since this gene encodes the reverse transcriptase for both viruses [5]. Since FIV shares many similarities to HIV it can be used as a model for HIV. Understanding FIV can also help us understand HIV because HIV seemingly jumped from primates to humans through hunting and there is speculation and evidence that FIV has also jumped speices in felines [5]. Also FIV has a similar progression of viral infection in cats as HIV infection has in humans and thus it can be studied as a model for HIV. FIV also shares a great genomic similarity to HIV especially in genes for the reverse transcriptase and protease [5]. The virus also has a very similar life cycle to that of HIV. Scientists can test new drugs and see the complete effects of immunodeficiency in cats and by doing so help cure HIV. Therefore understanding the genome, mutation rate and evolution of FIV can not only benefit Veterinary medicine, but human medicine as well.
References
1. The Cat Group. 2006. Policy Statement 3 Testing for Feline Immunodeficiency Virus (FIV). Journal of Feline Medicine and Surgery. 5: vi-viii.
2. Cornell Feline Health Center. 2002. Feline Immunodeficiency Virus. Cornell University College of Veterinary Medicine. Ithica, New York.
3. Gemeniano, Malou C., Sawai, Earl T., Sparger, Ellen E. 2004. Feline immunodeficiency virus Orf-A localizes to the nucleus and induces cell cycle arrest. Virology, 325: 167-`174.
4.Miyazawa, T., Tomonga, K., Kawaguchi, Y., Mikami, T. 1993. The genome of feline immunodeficiency virus. Virology. 134: 221-234.
5. Olmsted, Robert A., Hirsh, Vanessa M., Purcell, Robert H., Johnson, Phillip R. 1989. Nucleotide Sequence Analysis of Feline Immunodeficiency Virus: Genome Organization and Relationship to other Lentiviruses. Natural Academy of Sciences. 86: 8088-8092.
6. Pecan-Slattery, Jill. Troyer, Jennifer L. Johnson, Warren E., O’Brian, Stephen J. 2008. Evolution of feline immunodeficiency virus in Felidae: Implications for human heath and wildlife ecology. Veterinary immunology and immunopathology. 123: 32-44.
7. Slonczewski, Joan L., Foster, John W. Microbiology: An Evolving Science. 1st ed. New York: W.W. Norton and Company, Inc, 2009.
8. Tomonaga, Keizo, Shin, Yeon-Sli, Fukasawa, Masashi, Miyazawa, Takayuki, Adachi, Akio, Mikami, Takeshi. 1993. Feline immunodeficiency virus gene expression: analysis of the RNA splicing pattern and the monocistronic rev mRNA. Journal of General Virology. 74: 2409-2417.
9. Troyer, Jennifer L., VandeWoude, Sue, Pecon-Slattery, Jill, McIntosh, Carl, Franklin, Sam, Antunes, Agosinho, Johnson, Warren, O’Brian, Stephen. FIV cross-species transmission: An evolutionary pospective. Veterinary immunology and immunopathology. 123: 159-166.
Edited by Emily Nutt, student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.
