Fusarium venenatum
Classification
Fungi; Ascomycota; Sordariomycetes; Hypocreales; Nectriaceae
Species
|
NCBI: [1] |
Fusarium venenatum
Description and Significance
Fusarium venenatum is a filamentous, soil-dwelling, non-pathogenic fungi that is widespread in soils across the globe (King et al. 2018). It is closely related to Fusarium graminearum, a pathogenic fungus known to cause head blight on wheat which is also a producer of potent mycotoxins. When first cultured in the search for novel sources of mycoprotein, it was identified as F. graminearum, but further analysis of molecular phylogenetic, morphological, and mycotoxin data supported its reclassification as F. venenatum (O'Donnell et al. 1998). It is most well-known and studied as the fungus used as mycoprotein in the production of the food product known as Quorn (Wiebe 2002).
Life Cycle, Cell Structure, Metabolism
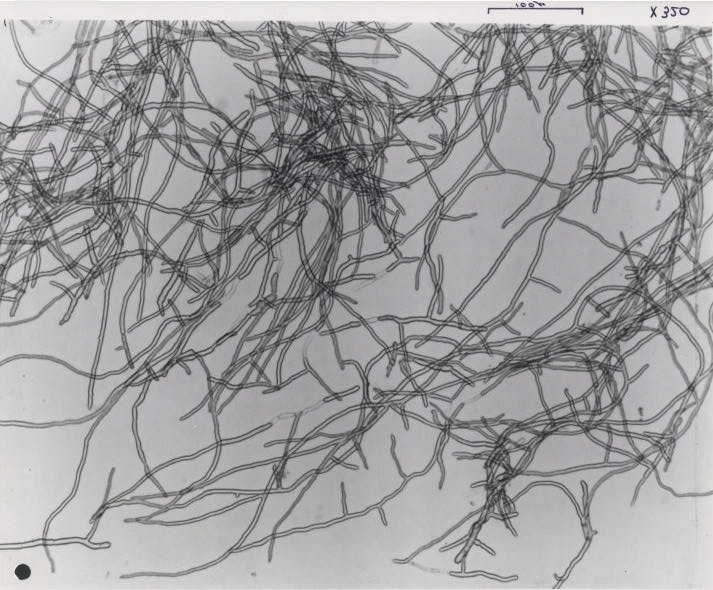
When grown in culture, F. venenatum produces multinucleate, multi- septate, banana-shaped spores known as macroconidia. The macroconidia are formed from uninucleate spore-producing cells known as phialides and are therefore homokaryotic. Mycelium of the fungi is coenocytic and shares the nuclear characteristics of the macroconidia (Trinci 1994). The fungus also produces two additional types of mitotic spores—microconidia produced from conidiophores, and clamydospres produced on and within hyphae (Dweba et al. 2016).
The generalized life cycle of Fusarium spp. has both sexual and asexual phases, both of which produce haploid mycelium. Following plasmogamy and karyogamy, outcorossed and selfed perithecium respectively produce recombinant and clonal meioticspores. These form haploid the mycelium which in turn form the three types of mitotic spores previously mentioned (Dweba et al. 2016).
Much research has been done on the industrial strains of F. venenatum used in the production of Quorn. Due to the fungus' metabolic processes and its ability to adapt to the large-scale fermentation processes necessary for its production as a food product, it has been examined as a potential producer of industrial enzymes such as trypsin and xylanase (Berka et al. 2004).
Genome Structure
F. venenatum has a genome 38,660,329 bp in length, which is assembled into four chromosomes. Current research leaves 37 gaps within the genome. The fungus' average gene length is 1388 bp, and the average centromere length of the species is 45 kbp.
DNA sequencing has definitively distinguished F. venenatum from the pathogenic F. graminearum through the presence of three genes in F. venenatum that are not found in F. graminearum—a transcription factor (FVRRES_13944), a cholinesterase (FVRRES_13945), and a negative transcriptional regulator.
Ecology and Pathogenesis
F. venenatum is a widely spread soil-borne fungus. Due to its relatedness to the pathogenic fungus F. graminearum, concerns have been raised about the ability of F. venenatum to cause disease in plants. Spray-inoculation of wheat heads and inoculation of wounds on tomatoes with both species showed little hyphal development of F. veneatum and no symptoms expressed in either the wheat head or tomato, whereas F. graminearum quickly colonized the tomato pericarp, and caused significant bleaching of the wheat head and aborted grain development (King et al. 2018).
F. venenatum is most significant as an industrial producer of mycoprotein. The species was discovered (but incorrectly classified as F. graminearum) in the 1960s as a result of a search for an organism that could convert starch into protein and provide an alternative source for protein production in the event of a global food shortage. The search for such an organism began in Britain when researchers sprayed a sports field with a starchy by-product of dairy production, hoping to select for wild organisms that could efficiently use starch as a substrate. The organisms cultured from the search of the field were unfortunately not able to grow efficiently in continuous culture (Finnigan et al. 2017). A broader search was initiated, and 3000 soil samples from around the globe were examined. Ultimately, F. venenatum was determined to be the best candidate for protein production and continuous cultivation in industrial contexts (Wiebe 2002).
References
Author
Page authored by Thom Plasse, student of Dr. Marc Orbach, University of Arizona .