Geomyces Destructans: Difference between revisions
Schmalingk (talk | contribs) |
Schmalingk (talk | contribs) |
||
| Line 44: | Line 44: | ||
<br /> A total of 9075 genes coding for proteins have been sequenced for Geomyces destructans in a genome of 29,706,106 base pairs to code for them. Liner mitochondrial DNA has also been sequences but little information is available on genetic specifics at this time. (NCBI)http://www.ncbi.nlm.nih.gov/pubmed/''. | <br /> A total of 9075 genes coding for proteins have been sequenced for Geomyces destructans in a genome of 29,706,106 base pairs to code for them. Liner mitochondrial DNA has also been sequences but little information is available on genetic specifics at this time. (NCBI)http://www.ncbi.nlm.nih.gov/pubmed/''. | ||
==Cell | ==Cell structure and metabolism== | ||
[[Image:Diphtheria.gif|thumb|200px|right|Morphology of <i>Corynebacterium Diphtheriae</i> [15]]] | |||
<b>Cell Exterior</b> | |||
Being Gram-positive, <i>C. diphtheriae </i>has a cell membrane and a lipid-rich murein layer outside the membrane. Like <i>M. tuberculosis</i>, another Actinomycete, <i>C. diphtheriae </i>has an arabinogalactan polymer which anchors a lipid rich domain to the murein layer. Corynomycolic acids, alpha-alkyl beta-hydroxy fatty acids, are produced through a Claisen-like condensation reaction between two fatty acyl chains. [1] | |||
Also in the exterior are the fimbriae (pili). Not much structural analysis has been done, however it is very likely that sortase-like proteins are produced to aid in anchoring the fimbriae and possibly polymerizing them. <i>C. diphtheriae </i>has six genes which code for putative sortases - five are localized on PAIs, none of which are found in closely related non-pathogenic corynebacteria (<i>C. glutamicum </i>and <i>C. efficens</i>). The absence of these genes in the non-pathogenic relatives suggests that those genes increase the pathogenicity of <i>C. diphtheriae </i>through the fimbriae, most likely by aiding the bacterium invade and adhere to host cells. The similarity to the sortase-related fimbrial system of Actinomyces, and the newness of the genes in the <i>C. diphtheria </i>genome, suggest that these genes were newly acquired, possibly the cause of the recent upsurge in outbreaks of Diphtheria. [1] | |||
<b>Metabolic processes </b> | |||
Pathways discovered for metabolism include the complete pentose phosphate pathway, glycolysis, and gluconeogenesis, based on the enzymes present in the cell. The citric acid cycle is present, however <i>C. diphtheriae </i>lacks succinyl-CoA synthetase and uses an enzyme transcribed by a homologue of the ''Clostridium kluyveri'' cat1 gene, which acts as a coenzyme A transferase rather than a synthetase. De novo amino acid biosynthesis is also present in <i>C. diphtheriae</i>, as well as both anaerobic and aerobic respiration pathways, though <i>C. diphtheriae </i>is primarily aerobic. Production of purines is complete, however of the pyrimidines, the cytidine production pathway lacks the final cytidine triphosphate synthetase, which is present in other related organisms such as <i>C. efficens </i>and <i>M. tuberculosis. </i>[1] | |||
<b>Specialized Iron Intake and Diphtheria Toxin</b> | |||
Diphtheria toxin is the most significant molecule produced by <i>C. diphtheriae</i>. The tox gene which codes for the Diphtheria toxin is located on the right-handed end of an integrated corynephage. <i>C. diptheriae</i> contains the protein DtxR, an iron-dependent negative-regulatory protein which prevents the tox gene from being transcribed under high iron conditions, as well as the high-energy iron uptake system of the bacterium. Because a host often reacts to infection by lowering iron levels, the bacteria have developed a mechanism which couples Diphtheria toxin gene expression with low iron levels. [1] | |||
To import iron, <i>C. diphtheriae </i>has seven different systems, two of which have been identified. The siderophore system creates ferrisiderophores, molecules specialized in binding to iron and bringing it into the cell for usage. [1] | |||
==Ecology== | ==Ecology== | ||
Revision as of 11:19, 3 May 2012
A Microbial Biorealm page on the genus Geomyces Destructans
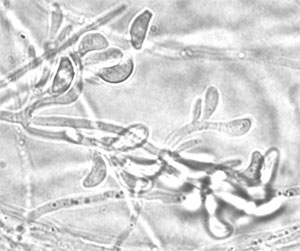
Classification
Kingdon: Fungi
Subkingdom: Dikarya
Phylum: Ascomycota
Subphylum: Pezizomycotina
Class: Leotiomycetes
Order: Helotiales
Family: Myxotrichaceae
Genus: Geomyces
Species: Geomyces destructans
Description and Significance
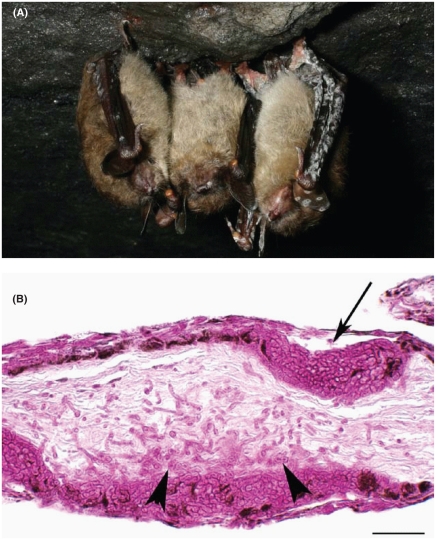
Geomices destructans is a fungus thought to be responsible for the large number of bat deaths in New England over the last several years. It first presented itself in the US in 2006 and then in Europe specifically France, in 2009. Until recently G. destructans was thought to be a normal resident inhabiting bats around the world. Further research links this to “white nose syndrome,” and even more recent research shows a 100% match between the DNA of G. destructans and the fungus found on the dead bats [1]. (Chaturvedi et al) Further study is needed on this subject; reachers do not seem to be sure if the fungus is stemming from pest control measures or how and why this has recently devastated bats around the world.
Since first observation near Albany, NY in 2006, white nose syndrome has decimated populations of cave-hibernating bats in the northeastern United States, with mortality rates of 75-95%. In 2009, the infection area extended from northeastern New Hampshire to southwestern Virginia and expanded into Tennessee and Canada in 2010 [2]. Outside of North America, G. destructans has also been observed in colonized bats across Europe. It is not yet obvious how the infection is influencing the bat populations in Europe [10]. Description and Significance
.
Genome Structure
A total of 9075 genes coding for proteins have been sequenced for Geomyces destructans in a genome of 29,706,106 base pairs to code for them. Liner mitochondrial DNA has also been sequences but little information is available on genetic specifics at this time. (NCBI)http://www.ncbi.nlm.nih.gov/pubmed/.
Cell structure and metabolism
Cell Exterior
Being Gram-positive, C. diphtheriae has a cell membrane and a lipid-rich murein layer outside the membrane. Like M. tuberculosis, another Actinomycete, C. diphtheriae has an arabinogalactan polymer which anchors a lipid rich domain to the murein layer. Corynomycolic acids, alpha-alkyl beta-hydroxy fatty acids, are produced through a Claisen-like condensation reaction between two fatty acyl chains. [1]
Also in the exterior are the fimbriae (pili). Not much structural analysis has been done, however it is very likely that sortase-like proteins are produced to aid in anchoring the fimbriae and possibly polymerizing them. C. diphtheriae has six genes which code for putative sortases - five are localized on PAIs, none of which are found in closely related non-pathogenic corynebacteria (C. glutamicum and C. efficens). The absence of these genes in the non-pathogenic relatives suggests that those genes increase the pathogenicity of C. diphtheriae through the fimbriae, most likely by aiding the bacterium invade and adhere to host cells. The similarity to the sortase-related fimbrial system of Actinomyces, and the newness of the genes in the C. diphtheria genome, suggest that these genes were newly acquired, possibly the cause of the recent upsurge in outbreaks of Diphtheria. [1]
Metabolic processes
Pathways discovered for metabolism include the complete pentose phosphate pathway, glycolysis, and gluconeogenesis, based on the enzymes present in the cell. The citric acid cycle is present, however C. diphtheriae lacks succinyl-CoA synthetase and uses an enzyme transcribed by a homologue of the Clostridium kluyveri cat1 gene, which acts as a coenzyme A transferase rather than a synthetase. De novo amino acid biosynthesis is also present in C. diphtheriae, as well as both anaerobic and aerobic respiration pathways, though C. diphtheriae is primarily aerobic. Production of purines is complete, however of the pyrimidines, the cytidine production pathway lacks the final cytidine triphosphate synthetase, which is present in other related organisms such as C. efficens and M. tuberculosis. [1]
Specialized Iron Intake and Diphtheria Toxin
Diphtheria toxin is the most significant molecule produced by C. diphtheriae. The tox gene which codes for the Diphtheria toxin is located on the right-handed end of an integrated corynephage. C. diptheriae contains the protein DtxR, an iron-dependent negative-regulatory protein which prevents the tox gene from being transcribed under high iron conditions, as well as the high-energy iron uptake system of the bacterium. Because a host often reacts to infection by lowering iron levels, the bacteria have developed a mechanism which couples Diphtheria toxin gene expression with low iron levels. [1]
To import iron, C. diphtheriae has seven different systems, two of which have been identified. The siderophore system creates ferrisiderophores, molecules specialized in binding to iron and bringing it into the cell for usage. [1]
Ecology
Due to the metabolic diversity in the genus Bacillus, bacilli are able to colonize a variety of habitats ranging from soil and insects to humans. Bacillus thuringiensis parasitizes insects, and is commercially used for pest control. Although the most well known of the bacilli are the pathogenic species, most Bacillus are saprophytes that make their living off of decaying matter. Still others, namely Bacillus subtilis, inhabit the rhizosphere, which is the interface between plant roots and the surrounding soil. The plants roots and associated biofilm can have a significant effect on the chemistry of the soil, creating a unique environment.
It has recently been shown that Bacillus subtilis engages in cannibalism. They use cannibalism as the easy way out in extreme cases. For survival in harsh environments, bacilli can form spores, but it is very costly to them energy-wise. An easier way is for the bacteria to produce antibiotics that destroy neighboring bacilli, so that their contents may be digested allowing for the survival of a few of the bacteria. Essentially, what they are doing is snacking on their fellow bacilli, to tide them over, hoping for the environment to pick back up.
Pathology
Bacilli cause an array of infections from ear infections to meningitis, and urinary tract infections to septicemia. Mostly they occur as secondary infections in immunodeficient hosts or otherwise compromised hosts. They may exacerbate previous infection by producing tissue-damaging toxins or metabolites that interfere with treatment.
The most well known disease caused by bacilli is anthrax, caused by Bacillus anthracis. Anthrax has a long history with humans. It has been suggested that the fifth and sixth plagues of Egypt recorded in the Bible (the fifth attacking animals, the sixth, known as the plague of the boils, attacking humans). In the 1600s anthrax was known as the "Black bane" and killed over 60,000 cows. Anthrax has more recently been brought to our attention as a possible method for bioterrorism. The recent anthrax mailings have brought acute public attention to the issue and sparked extensive research into the devastating disease.
Anthrax is primarily a disease of herbivores who acquire the bacterium by eating plants with dust that contains anthrax spores. Humans contract the disease in three different ways. Cutaneous anthrax occurs when a human comes into contact with the spores form dust particles or a contaminated animal or carcass through a cut or abrasion. Cutaneous anthrax accounts for 95% of anthrax cases worldwide. During a 2-3 day incubation period the spores germinate, vegetative cells multiply, and a papule develops. Over the following days the papule ulcerates, dries and blackens to form the characteristic eschar. The process is painless unless infected with another pathogen.
Gastrointestinal anthrax is contracted by ingesting contaminated meat. It occurs in the intestinal mucosa when the organisms invade the mucosa through a preexisting lesions. It progresses the same way as cutaneous anthrax. Although it is extremely rare in developed countries, it has a very high mortality rate.
Pulmonary anthrax is the result of inhaled spores that are transported to the lymph nodes where they germinate and multiply. They are then taken into the blood stream and lymphatics culminating in systemic arthritis which is usually fatal.
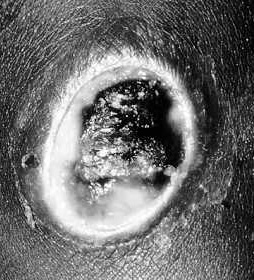
Phages
Due to the danger of anthrax being used in biological weapons, research has been put into other methods, besides the highly controversial vaccine, to defend against the deadly disease. A recently discovered bacteriophage, the gamma phage, attacks Bacillus anthracis, and researches are optimistic about its clinical application. The bacteriophage is highly selective, and is extremely effective in lysing B. anthracis cells, while ignoring those of its closely related counterparts B. cereus and B. thuringiensis. The gamma phage has been over 80% effective in treating infected mice that were in the late stages of the disease, essentially rescuing them from almost certain death. There is the obvious concern that anthrax will develop strains that are immune to this treatment, and we will be right back where we started. Researchers say that this is unlikely because the only way to evade this predator would be a mutational change in cell wall structure to prevent the virus from binding, and this would kill the bacterium.
Medicine
Despite the pathogenic capabilities of some bacilli, many other species are used in medical and pharmaceutical processes. These take advantage of the bacteria's ability to synthesize certain proteins and antibiotics. Bacitracin and plymixin, two ingredients in Neosporin, are products of bacilli. Also, innocuous Bacillus microbes are useful for studying the virulent bacillus species that are closely related. B. subtilis has multiple carbohydrate pathways, representing the variety of carbohydrates found in the soil.
References
Innovation in Europe: The decoding of the Bacillus subtilis genome
University of Texas Medical Branch: Bacillus
University of Wisconsin-Madison: Bacillus anthracis and anthrax
Wipat, Anil & Colin R. Hardwood. 1999. The Bacillus subtilis genome sequence: the molecular blueprint of a soil bacterium. FEMS Microbiology Ecology, Vol. 28: 1-9.

